Dissecting Nucleosome Function with a Comprehensive Histone H2A and H2B Mutant Library
- PMID: 29038170
- PMCID: PMC5714483
- DOI: 10.1534/g3.117.300252
Dissecting Nucleosome Function with a Comprehensive Histone H2A and H2B Mutant Library
Abstract
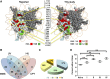
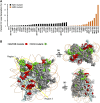
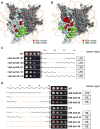
Using a comprehensive library of histone H2A and H2B mutants, we assessed the biological function of each amino acid residue involved in various stress conditions including exposure to different DNA damage-inducing reagents, different growth temperatures, and other chemicals. H2B N- and H2A C-termini were critical for maintaining nucleosome function and mutations in these regions led to pleiotropic phenotypes. Additionally, two screens were performed using this library, monitoring heterochromatin gene silencing and genome stability, to identify residues that could compromise normal function when mutated. Many distinctive regions within the nucleosome were revealed. Furthermore, we used the barcode sequencing (bar-seq) method to profile the mutant composition of many libraries in one high-throughput sequencing experiment, greatly reducing the labor and increasing the capacity. This study not only demonstrates the applications of the versatile histone library, but also reveals many previously unknown functions of histone H2A and H2B.
Keywords: DNA damage; heterochromatin gene silencing; histone; post-translational modification.
Copyright © 2017 Jiang et al.
Figures


Similar articles
-
Residues in the Nucleosome Acidic Patch Regulate Histone Occupancy and Are Important for FACT Binding in Saccharomyces cerevisiae.Genetics. 2017 Jul;206(3):1339-1348. doi: 10.1534/genetics.117.201939. Epub 2017 May 3. Genetics. 2017. PMID: 28468903 Free PMC article.
-
Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi-Snf in yeast.EMBO J. 1999 Jan 4;18(1):229-40. doi: 10.1093/emboj/18.1.229. EMBO J. 1999. PMID: 9878065 Free PMC article.
-
Histone H2A subtypes associate interchangeably in vivo with histone H2B subtypes.Proc Natl Acad Sci U S A. 1982 Dec;79(24):7814-8. doi: 10.1073/pnas.79.24.7814. Proc Natl Acad Sci U S A. 1982. PMID: 6760203 Free PMC article.
-
The role of histone H2A and H2B post-translational modifications in transcription: a genomic perspective.Biochim Biophys Acta. 2009 Jan;1789(1):37-44. doi: 10.1016/j.bbagrm.2008.07.001. Epub 2008 Jul 14. Biochim Biophys Acta. 2009. PMID: 18675384 Review.
-
Histone H2A variants H2AX and H2AZ.Curr Opin Genet Dev. 2002 Apr;12(2):162-9. doi: 10.1016/s0959-437x(02)00282-4. Curr Opin Genet Dev. 2002. PMID: 11893489 Review.
Cited by
-
Control of Gene Expression via the Yeast CWI Pathway.Int J Mol Sci. 2022 Feb 4;23(3):1791. doi: 10.3390/ijms23031791. Int J Mol Sci. 2022. PMID: 35163713 Free PMC article. Review.
-
Histone Modifications as an Intersection Between Diet and Longevity.Front Genet. 2019 Mar 12;10:192. doi: 10.3389/fgene.2019.00192. eCollection 2019. Front Genet. 2019. PMID: 30915107 Free PMC article. Review.
-
Structure and function of the Orc1 BAH-nucleosome complex.Nat Commun. 2019 Jul 1;10(1):2894. doi: 10.1038/s41467-019-10609-y. Nat Commun. 2019. PMID: 31263106 Free PMC article.
-
Retrotransposon targeting to RNA polymerase III-transcribed genes.Mob DNA. 2018 Apr 23;9:14. doi: 10.1186/s13100-018-0119-2. eCollection 2018. Mob DNA. 2018. PMID: 29713390 Free PMC article. Review.
-
Comprehensive nucleosome interactome screen establishes fundamental principles of nucleosome binding.Nucleic Acids Res. 2020 Sep 25;48(17):9415-9432. doi: 10.1093/nar/gkaa544. Nucleic Acids Res. 2020. PMID: 32658293 Free PMC article.
References
-
- Aguilera J., Randez-Gil F., Prieto J. A., 2007. Cold response in Saccharomyces cerevisiae: new functions for old mechanisms. FEMS Microbiol. Rev. 31: 327–341. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
