A CTG Clade Candida Yeast Genetically Engineered for the Genotype-Phenotype Characterization of Azole Antifungal Resistance in Human-Pathogenic Yeasts
- PMID: 29038279
- PMCID: PMC5740319
- DOI: 10.1128/AAC.01483-17
A CTG Clade Candida Yeast Genetically Engineered for the Genotype-Phenotype Characterization of Azole Antifungal Resistance in Human-Pathogenic Yeasts
Abstract
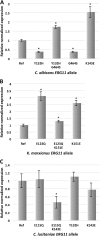
A strain of the opportunistic pathogenic yeast Candida lusitaniae was genetically modified for use as a cellular model for assessing by allele replacement the impact of lanosterol C14α-demethylase ERG11 mutations on azole resistance. Candida lusitaniae was chosen because it is susceptible to azole antifungals, it belongs to the CTG clade of yeast, which includes most of the Candida species pathogenic for humans, and it is haploid and easily amenable to genetic transformation and molecular modeling. In this work, allelic replacement is targeted at the ERG11 locus by the reconstitution of a functional auxotrophic marker in the 3' intergenic region of ERG11 Homologous and heterologous ERG11 alleles are expressed from the resident ERG11 promoter of C. lusitaniae, allowing accurate comparison of the phenotypic change in azole susceptibility. As a proof of concept, we successfully expressed in C. lusitaniae different ERG11 alleles, either bearing or not bearing mutations retrieved from a clinical context, from two phylogenetically distant yeasts, C. albicans and Kluyveromyces marxianusCandida lusitaniae constitutes a high-fidelity expression system, giving specific Erg11p-dependent fluconazole MICs very close to those observed with the ERG11 donor strain. This work led us to characterize the phenotypic effect of two kinds of mutation: mutation conferring decreased fluconazole susceptibility in a species-specific manner and mutation conferring fluconazole resistance in several yeast species. In particular, a missense mutation affecting amino acid K143 of Erg11p in Candida species, and the equivalent position K151 in K. marxianus, plays a critical role in fluconazole resistance.
Keywords: Candida; Candida lusitaniae; ERG11 mutation; Kluyveromyces; fluconazole resistance; heterologous expression.
Copyright © 2017 American Society for Microbiology.
Figures



References
-
- des Champs-Bro B, Leroy-Cotteau A, Mazingue F, Pasquier F, François N, Corm S, Lemaitre L, Poulain D, Yakoub-Agha I, Alfandari S, Sendid B. 2011. Invasive fungal infections: epidemiology and analysis of antifungal prescriptions in onco-haematology. J Clin Pharm Ther 36:152–160. doi:10.1111/j.1365-2710.2010.01166.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

