The small GTPase RSG1 controls a final step in primary cilia initiation
- PMID: 29038301
- PMCID: PMC5748968
- DOI: 10.1083/jcb.201604048
The small GTPase RSG1 controls a final step in primary cilia initiation
Abstract
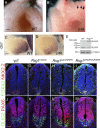
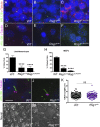
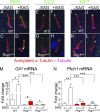
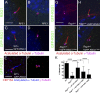
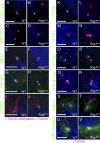
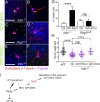
Primary cilia, which are essential for normal development and tissue homeostasis, are extensions of the mother centriole, but the mechanisms that remodel the centriole to promote cilia initiation are poorly understood. Here we show that mouse embryos that lack the small guanosine triphosphatase RSG1 die at embryonic day 12.5, with developmental abnormalities characteristic of decreased cilia-dependent Hedgehog signaling. Rsg1 mutant embryos have fewer primary cilia than wild-type embryos, but the cilia that form are of normal length and traffic Hedgehog pathway proteins within the cilium correctly. Rsg1 mother centrioles recruit proteins required for cilia initiation and dock onto ciliary vesicles, but axonemal microtubules fail to elongate normally. RSG1 localizes to the mother centriole in a process that depends on tau tubulin kinase 2 (TTBK2), the CPLANE complex protein Inturned (INTU), and its own GTPase activity. The data suggest a specific role for RSG1 in the final maturation of the mother centriole and ciliary vesicle that allows extension of the ciliary axoneme.
© 2018 Agbu et al.
Figures


Similar articles
-
Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis.J Cell Biol. 2012 Dec 24;199(7):1083-101. doi: 10.1083/jcb.201202126. Epub 2012 Dec 17. J Cell Biol. 2012. PMID: 23253480 Free PMC article.
-
Rab34 small GTPase is required for Hedgehog signaling and an early step of ciliary vesicle formation in mouse.J Cell Sci. 2018 Nov 8;131(21):jcs213710. doi: 10.1242/jcs.213710. J Cell Sci. 2018. PMID: 30301781 Free PMC article.
-
CEP162 is an axoneme-recognition protein promoting ciliary transition zone assembly at the cilia base.Nat Cell Biol. 2013 Jun;15(6):591-601. doi: 10.1038/ncb2739. Epub 2013 May 5. Nat Cell Biol. 2013. PMID: 23644468 Free PMC article.
-
CPLANE Complex and Ciliopathies.Biomolecules. 2022 Jun 17;12(6):847. doi: 10.3390/biom12060847. Biomolecules. 2022. PMID: 35740972 Free PMC article. Review.
-
The Ciliary Transition Zone: Finding the Pieces and Assembling the Gate.Mol Cells. 2017 Apr;40(4):243-253. doi: 10.14348/molcells.2017.0054. Epub 2017 Apr 12. Mol Cells. 2017. PMID: 28401750 Free PMC article. Review.
Cited by
-
Cilia proteins getting to work - how do they commute from the cytoplasm to the base of cilia?J Cell Sci. 2022 Sep 1;135(17):jcs259444. doi: 10.1242/jcs.259444. Epub 2022 Sep 8. J Cell Sci. 2022. PMID: 36073764 Free PMC article. Review.
-
A WDR35-dependent coat protein complex transports ciliary membrane cargo vesicles to cilia.Elife. 2021 Nov 4;10:e69786. doi: 10.7554/eLife.69786. Elife. 2021. PMID: 34734804 Free PMC article.
-
RSG1 is required for cilia-dependent neural tube closure.Genesis. 2024 Jun;62(3):e23602. doi: 10.1002/dvg.23602. Genesis. 2024. PMID: 38721990 Free PMC article.
-
CPLANE protein INTU regulates growth and patterning of the mouse lungs through cilia-dependent Hh signaling.Dev Biol. 2024 Nov;515:92-101. doi: 10.1016/j.ydbio.2024.07.006. Epub 2024 Jul 17. Dev Biol. 2024. PMID: 39029571
-
The regulation of cilium assembly and disassembly in development and disease.Development. 2018 Sep 17;145(18):dev151407. doi: 10.1242/dev.151407. Development. 2018. PMID: 30224385 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

