The contribution of Saccharomyces cerevisiae replicative age to the variations in the levels of Trx2p, Pdr5p, Can1p and Idh isoforms
- PMID: 29038504
- PMCID: PMC5643315
- DOI: 10.1038/s41598-017-13576-w
The contribution of Saccharomyces cerevisiae replicative age to the variations in the levels of Trx2p, Pdr5p, Can1p and Idh isoforms
Abstract
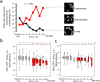
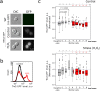
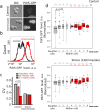
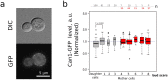
Asymmetrical division can be a reason for microbial populations heterogeneity. In particular, budding yeast daughter cells are more vulnerable to stresses than the mothers. It was suggested that yeast mother cells could also differ from each other depending on their replicative age. To test this, we measured the levels of Idh1-GFP, Idh2-GFP, Trx2-GFP, Pdr5-GFP and Can1-GFP proteins in cells of the few first, most represented, age cohorts. Pdr5p and Can1p were selected because of the pronounced mother-bud asymmetry for these proteins distributions, Trx2p as indicator of oxidative stress. Isocitrate dehydrogenase subunits Idh1p and Idh2p were assessed because their levels are regulated by mitochondria. We found a small negative correlation between yeast replicative age and Idh1-GFP or Idh2-GFP but not Trx2-GFP levels. Mitochondrial network fragmentation was also confirmed as an early event of replicative aging. No significant difference in the membrane proteins levels Pdr5p and Can1p was found. Moreover, the elder mother cells showed lower coefficient of variation for Pdr5p levels compared to the younger ones and the daughters. Our data suggest that the levels of stress-response proteins Pdr5p and Trx2p in the mother cells are stable during the first few cell cycles regardless of their mother-bud asymmetry.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Mutations of charged amino acids at the cytoplasmic end of transmembrane helix 2 affect transport activity of the budding yeast multidrug resistance protein Pdr5p.FEMS Yeast Res. 2016 Jun;16(4):fow031. doi: 10.1093/femsyr/fow031. Epub 2016 Apr 19. FEMS Yeast Res. 2016. PMID: 27189366 Free PMC article.
-
Involvement of Saccharomyces cerevisiae Pdr5p ATP-binding cassette transporter in calcium homeostasis.Biosci Biotechnol Biochem. 2005 Apr;69(4):857-60. doi: 10.1271/bbb.69.857. Biosci Biotechnol Biochem. 2005. PMID: 15849433
-
Sir2-dependent daughter-to-mother transport of the damaged proteins in yeast is required to prevent high stress sensitivity of the daughters.Cell Cycle. 2010 Nov 15;9(22):4501-5. doi: 10.4161/cc.9.22.13683. Cell Cycle. 2010. PMID: 21084861
-
A mother's sacrifice: what is she keeping for herself?Curr Opin Cell Biol. 2008 Dec;20(6):723-8. doi: 10.1016/j.ceb.2008.09.004. Epub 2008 Oct 23. Curr Opin Cell Biol. 2008. PMID: 18848886 Free PMC article. Review.
-
Role of Mitochondrial DNA in Yeast Replicative Aging.Biochemistry (Mosc). 2023 Dec;88(12):1997-2006. doi: 10.1134/S0006297923120040. Biochemistry (Mosc). 2023. PMID: 38462446 Review.
Cited by
-
Replicative Aging in Pathogenic Fungi.J Fungi (Basel). 2020 Dec 25;7(1):6. doi: 10.3390/jof7010006. J Fungi (Basel). 2020. PMID: 33375605 Free PMC article. Review.
-
With age comes resilience: how mitochondrial modulation drives age-associated fluconazole tolerance in Cryptococcus neoformans.mBio. 2024 Sep 11;15(9):e0184724. doi: 10.1128/mbio.01847-24. Epub 2024 Aug 13. mBio. 2024. PMID: 39136442 Free PMC article.
-
Enhanced Efflux Pump Activity in Old Candida glabrata Cells.Antimicrob Agents Chemother. 2018 Feb 23;62(3):e02227-17. doi: 10.1128/AAC.02227-17. Print 2018 Mar. Antimicrob Agents Chemother. 2018. PMID: 29311061 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

