TRPA1-FGFR2 binding event is a regulatory oncogenic driver modulated by miRNA-142-3p
- PMID: 29038531
- PMCID: PMC5643494
- DOI: 10.1038/s41467-017-00983-w
TRPA1-FGFR2 binding event is a regulatory oncogenic driver modulated by miRNA-142-3p
Abstract
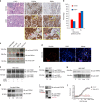
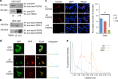
Recent evidence suggests that the ion channel TRPA1 is implicated in lung adenocarcinoma (LUAD), where its role and mechanism of action remain unknown. We have previously established that the membrane receptor FGFR2 drives LUAD progression through aberrant protein-protein interactions mediated via its C-terminal proline-rich motif. Here we report that the N-terminal ankyrin repeats of TRPA1 directly bind to the C-terminal proline-rich motif of FGFR2 inducing the constitutive activation of the receptor, thereby prompting LUAD progression and metastasis. Furthermore, we show that upon metastasis to the brain, TRPA1 gets depleted, an effect triggered by the transfer of TRPA1-targeting exosomal microRNA (miRNA-142-3p) from brain astrocytes to cancer cells. This downregulation, in turn, inhibits TRPA1-mediated activation of FGFR2, hindering the metastatic process. Our study reveals a direct binding event and characterizes the role of TRPA1 ankyrin repeats in regulating FGFR2-driven oncogenic process; a mechanism that is hindered by miRNA-142-3p.TRPA1 has been reported to contribute lung cancer adenocarcinoma (LUAD), but the mechanisms are unclear. Here the authors propose that TRPA1/FGFR2 interaction is functional in LUAD and show that astrocytes oppose brain metastasis by mediating the downregulation of TRPA1 through exosome-delivered miRNA-142-3p.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

