Anti-Infective Effect of Adhesive Probiotic Lactobacillus in Fish is Correlated With Their Spatial Distribution in the Intestinal Tissue
- PMID: 29038557
- PMCID: PMC5643340
- DOI: 10.1038/s41598-017-13466-1
Anti-Infective Effect of Adhesive Probiotic Lactobacillus in Fish is Correlated With Their Spatial Distribution in the Intestinal Tissue
Abstract
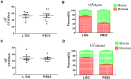
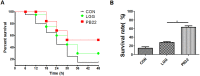
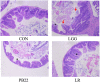
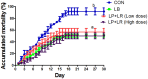
In this study, we tested the distribution of 49 Lactobacillus strains in the mucus and mucosa of the intestine tissue of zebrafish. We observed a progressive change in the spatial distribution of Lactobacillus strains, and suggested a division of the strains into three classes: mucus type (>70% in mucus), mucosa type (>70% in mucosa) and hybrid type (others). The hybrid type strains were more efficient in protection of zebrafish against Aeromonas hydrophila infection. Three strains representing different distribution types (JCM1149, CGMCC1.2028, and JCM 20300) were selected. The mucosa type strain JCM1149 induced higher intestinal expression of inflammatory cytokines and Hsp70 than the other strains. Furthermore, we used L. rhamnosus GG and its mutant (PB22) lacking SpaCBA pili to investigate the influence of pili on spatial distribution. LGG showed a mucosa type distribution, while PB22 revealed a hybrid distribution and the disease protection was accordingly improved. The different protection ability between LGG and PB22 did not involve the intestinal microbiota, however, LGG induced injury to the mucosa of zebrafish. Collectively, the disease protection activity of Lactobacillus in zebrafish is correlated with their spatial distribution in the intestinal tissue, with strains showing a balanced distribution (hybrid type) more efficient in protection.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Abrupt suspension of probiotics administration may increase host pathogen susceptibility by inducing gut dysbiosis.Sci Rep. 2016 Mar 17;6:23214. doi: 10.1038/srep23214. Sci Rep. 2016. PMID: 26983596 Free PMC article.
-
Protection mechanism of probiotic combination against human pathogens: in vitro adhesion to human intestinal mucus.Asia Pac J Clin Nutr. 2006;15(4):570-5. Asia Pac J Clin Nutr. 2006. PMID: 17077078 Review.
-
Probiotic strains and their combination inhibit in vitro adhesion of pathogens to pig intestinal mucosa.Curr Microbiol. 2007 Sep;55(3):260-5. doi: 10.1007/s00284-007-0144-8. Epub 2007 Jul 25. Curr Microbiol. 2007. PMID: 17657533
-
Development of new probiotics by strain combinations: is it possible to improve the adhesion to intestinal mucus?J Dairy Sci. 2007 Jun;90(6):2710-6. doi: 10.3168/jds.2006-456. J Dairy Sci. 2007. PMID: 17517710
-
Probiotics in foods not containing milk or milk constituents, with special reference to Lactobacillus plantarum 299v.Am J Clin Nutr. 2001 Feb;73(2 Suppl):380S-385S. doi: 10.1093/ajcn/73.2.380s. Am J Clin Nutr. 2001. PMID: 11157345 Review.
Cited by
-
Zebrafish: an efficient vertebrate model for understanding role of gut microbiota.Mol Med. 2022 Dec 23;28(1):161. doi: 10.1186/s10020-022-00579-1. Mol Med. 2022. PMID: 36564702 Free PMC article. Review.
-
Characterisation of Lactic Acid Bacteria from Dengke Naniura of Common Carp (Cyprinus carpio) with α-Glucosidase Inhibitory Activity.Open Access Maced J Med Sci. 2019 Nov 14;7(22):3794-3798. doi: 10.3889/oamjms.2019.506. eCollection 2019 Nov 30. Open Access Maced J Med Sci. 2019. PMID: 32127978 Free PMC article.
-
Commensal Microbiota Regulate Vertebrate Innate Immunity-Insights From the Zebrafish.Front Immunol. 2019 Sep 6;10:2100. doi: 10.3389/fimmu.2019.02100. eCollection 2019. Front Immunol. 2019. PMID: 31555292 Free PMC article. Review.
-
The Microbiota and Gut-Related Disorders: Insights from Animal Models.Cells. 2020 Nov 2;9(11):2401. doi: 10.3390/cells9112401. Cells. 2020. PMID: 33147801 Free PMC article. Review.
-
Lacticaseibacillus casei ATCC 393 Cannot Colonize the Gastrointestinal Tract of Crucian Carp.Microorganisms. 2021 Dec 9;9(12):2547. doi: 10.3390/microorganisms9122547. Microorganisms. 2021. PMID: 34946147 Free PMC article.
References
-
- Zhou Z, et al. Identification of highly-adhesive gut Lactobacillus strains in zebrafish (Danio rerio) by partial rpoB gene sequence analysis. Aquaculture. 2012;370:150–157. doi: 10.1016/j.aquaculture.2012.10.018. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

