RNA sequencing analysis of activated macrophages treated with the anti-HIV ABX464 in intestinal inflammation
- PMID: 29039845
- PMCID: PMC5644369
- DOI: 10.1038/sdata.2017.150
RNA sequencing analysis of activated macrophages treated with the anti-HIV ABX464 in intestinal inflammation
Abstract
RNA-Seq enables the generation of extensive transcriptome information providing the capability to characterize transcripts (including alternative isoforms and polymorphism), to quantify expression and to identify differential regulation in a single experiment. To reveal the capacity of new anti-HIV ABX464 candidate in modulating the expression of genes, datasets were generated and validated using RNA-seq approach. This comprehensive dataset will be useful to deepen the comprehensive understanding of the progression of human immunodeficiency virus (HIV) associated with mucosal damage in the gastrointestinal (GI) tract and subsequent inflammation, providing an opportunity to generate new therapies, diagnoses, and preventive strategies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
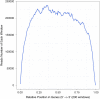
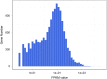

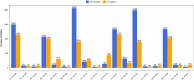

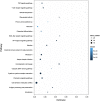
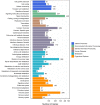
Dataset use reported in
- doi: 10.1038/s41598-017-04071-3
References
Data Citations
-
- Chebli K., Papon L., Hahne M., Manchon L., Tazi J. 2017. Figshare. https://dx.doi.org/10.6084/m9.figshare.4811035 - DOI - PubMed
-
- 2017. NCBI Gene Expression Omnibus. GSE97062
-
- Chebli K., Papon L., Hahne M., Manchon L., Tazi J. 2017. Figshare. https://dx.doi.org/10.6084/m9.figshare.4757974 - DOI - PubMed
-
- Chebli K., Papon L., Hahne M., Manchon L., Tazi J. 2017. Figshare. https://dx.doi.org/10.6084/m9.figshare.4810825 - DOI - PubMed
-
- Chebli K., Papon L., Hahne M., Manchon L., Tazi J. 2017. Figshare. https://dx.doi.org/10.6084/m9.figshare.4811077 - DOI - PubMed
References
-
- Chebli K. et al. The anti-HIV candidate ABX464 dampens intestinal inflammation by triggering IL22 production in activated macrophages. Scientific Reports 7, 4860 https://dx.doi.org/doi:10.1038/s41598-017-04071-3 (2017). - PMC - PubMed
-
- Mortazavi A. et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 5, 621–628 (2008). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

