Machine learning accelerates MD-based binding pose prediction between ligands and proteins
- PMID: 29040432
- PMCID: PMC6030886
- DOI: 10.1093/bioinformatics/btx638
Machine learning accelerates MD-based binding pose prediction between ligands and proteins
Abstract
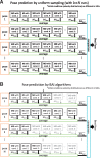
Motivation: Fast and accurate prediction of protein-ligand binding structures is indispensable for structure-based drug design and accurate estimation of binding free energy of drug candidate molecules in drug discovery. Recently, accurate pose prediction methods based on short Molecular Dynamics (MD) simulations, such as MM-PBSA and MM-GBSA, among generated docking poses have been used. Since molecular structures obtained from MD simulation depend on the initial condition, taking the average over different initial conditions leads to better accuracy. Prediction accuracy of protein-ligand binding poses can be improved with multiple runs at different initial velocity.
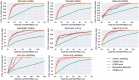
Results: This paper shows that a machine learning method, called Best Arm Identification, can optimally control the number of MD runs for each binding pose. It allows us to identify a correct binding pose with a minimum number of total runs. Our experiment using three proteins and eight inhibitors showed that the computational cost can be reduced substantially without sacrificing accuracy. This method can be applied for controlling all kinds of molecular simulations to obtain best results under restricted computational resources.
Availability and implementation: Code and data are available on GitHub at https://github.com/tsudalab/bpbi.
Contact: terayama@cbms.k.u-tokyo.ac.jp or tsuda@k.u-tokyo.ac.jp.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2017. Published by Oxford University Press.
Figures




Similar articles
-
Machine learning in computational docking.Artif Intell Med. 2015 Mar;63(3):135-52. doi: 10.1016/j.artmed.2015.02.002. Epub 2015 Feb 16. Artif Intell Med. 2015. PMID: 25724101
-
Exploring the Stability of Ligand Binding Modes to Proteins by Molecular Dynamics Simulations: A Cross-docking Study.J Chem Inf Model. 2017 Oct 23;57(10):2514-2522. doi: 10.1021/acs.jcim.7b00412. Epub 2017 Sep 29. J Chem Inf Model. 2017. PMID: 28902511
-
Exploring the stability of ligand binding modes to proteins by molecular dynamics simulations.J Comput Aided Mol Des. 2017 Feb;31(2):201-211. doi: 10.1007/s10822-016-0005-2. Epub 2017 Jan 10. J Comput Aided Mol Des. 2017. PMID: 28074360
-
Post processing of protein-compound docking for fragment-based drug discovery (FBDD): in-silico structure-based drug screening and ligand-binding pose prediction.Curr Top Med Chem. 2010;10(6):680-94. doi: 10.2174/156802610791111452. Curr Top Med Chem. 2010. PMID: 20337587 Review.
-
Cryptic binding sites on proteins: definition, detection, and druggability.Curr Opin Chem Biol. 2018 Jun;44:1-8. doi: 10.1016/j.cbpa.2018.05.003. Epub 2018 May 23. Curr Opin Chem Biol. 2018. PMID: 29800865 Free PMC article. Review.
Cited by
-
In-Silico Approaches for the Screening and Discovery of Broad-Spectrum Marine Natural Product Antiviral Agents Against Coronaviruses.Infect Drug Resist. 2023 Apr 19;16:2321-2338. doi: 10.2147/IDR.S395203. eCollection 2023. Infect Drug Resist. 2023. PMID: 37155475 Free PMC article. Review.
-
Fragment-centric topographic mapping method guides the understanding of ABCG2-inhibitor interactions.RSC Adv. 2019 Mar 8;9(14):7757-7766. doi: 10.1039/c8ra09789e. eCollection 2019 Mar 6. RSC Adv. 2019. PMID: 35521159 Free PMC article.
-
DROIDS 3.0-Detecting Genetic and Drug Class Variant Impact on Conserved Protein Binding Dynamics.Biophys J. 2020 Feb 4;118(3):541-551. doi: 10.1016/j.bpj.2019.12.008. Epub 2019 Dec 18. Biophys J. 2020. PMID: 31928763 Free PMC article.
-
From Byte to Bench to Bedside: Molecular Dynamics Simulations and Drug Discovery.ArXiv [Preprint]. 2023 Nov 28:arXiv:2311.16946v1. ArXiv. 2023. Update in: BMC Biol. 2023 Dec 29;21(1):299. doi: 10.1186/s12915-023-01791-z. PMID: 38076508 Free PMC article. Updated. Preprint.
-
In silico Prediction, Characterization, Molecular Docking, and Dynamic Studies on Fungal SDRs as Novel Targets for Searching Potential Fungicides Against Fusarium Wilt in Tomato.Front Pharmacol. 2018 Oct 22;9:1038. doi: 10.3389/fphar.2018.01038. eCollection 2018. Front Pharmacol. 2018. PMID: 30405403 Free PMC article.
References
-
- Agrawal S., Goyal N. (2012) Analysis of Thompson sampling for the multi-armed bandit problem. In: Conference on Learning Theory, pp. 39.1–39.26.
-
- Åqvist J. et al. (1994) A new method for predicting binding affinity in computer-aided drug design. Protein Eng., 7, 385–391. - PubMed
-
- Audibert J.-Y., Bubeck S. (2010) Best arm identification in multi-armed bandits. In: Conference on Learning Theory, p. 13. Haifa, Israel.
-
- Auer P. et al. (2002) Finite-time analysis of the multiarmed bandit problem. Mach. Learn., 47, 235–256.

