The MRPP1/MRPP2 complex is a tRNA-maturation platform in human mitochondria
- PMID: 29040705
- PMCID: PMC5716156
- DOI: 10.1093/nar/gkx902
The MRPP1/MRPP2 complex is a tRNA-maturation platform in human mitochondria
Abstract
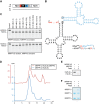
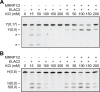
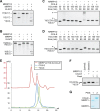
Mitochondrial polycistronic transcripts are extensively processed to give rise to functional mRNAs, rRNAs and tRNAs; starting with the release of tRNA elements through 5'-processing by RNase P (MRPP1/2/3-complex) and 3'-processing by RNase Z (ELAC2). Here, we show using in vitro experiments that MRPP1/2 is not only a component of the mitochondrial RNase P but that it retains the tRNA product from the 5'-processing step and significantly enhances the efficiency of ELAC2-catalyzed 3'-processing for 17 of the 22 tRNAs encoded in the human mitochondrial genome. Furthermore, MRPP1/2 retains the tRNA product after ELAC2 processing and presents the nascent tRNA to the mitochondrial CCA-adding enzyme. Thus, in addition to being an essential component of the RNase P reaction, MRPP1/2 serves as a processing platform for several down-stream tRNA maturation steps in human mitochondria. These findings are of fundamental importance for our molecular understanding of disease-related mutations in MRPP1/2, ELAC2 and mitochondrial tRNA genes.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Hällberg B.M., Larsson N.G.. Making proteins in the powerhouse. Cell Metab. 2014; 20:226–240. - PubMed
-
- Kauppila T.E., Kauppila J.H., Larsson N.G.. Mammalian mitochondria and aging: an update. Cell Metab. 2017; 25:57–71. - PubMed
-
- Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F. et al. Sequence and organization of the human mitochondrial genome. Nature. 1981; 290:457–465. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

