An analysis of 67 RNA-seq datasets from various tissues at different stages of a model insect, Manduca sexta
- PMID: 29041902
- PMCID: PMC5645894
- DOI: 10.1186/s12864-017-4147-y
An analysis of 67 RNA-seq datasets from various tissues at different stages of a model insect, Manduca sexta
Abstract
Background: Manduca sexta is a large lepidopteran insect widely used as a model to study biochemistry of insect physiological processes. As a part of its genome project, over 50 cDNA libraries have been analyzed to profile gene expression in different tissues and life stages. While the RNA-seq data were used to study genes related to cuticle structure, chitin metabolism and immunity, a vast amount of the information has not yet been mined for understanding the basic molecular biology of this model insect. In fact, the basic features of these data, such as composition of the RNA-seq reads and lists of library-correlated genes, are unclear. From an extended view of all insects, clear-cut tempospatial expression data are rarely seen in the largest group of animals including Drosophila and mosquitoes, mainly due to their small sizes.
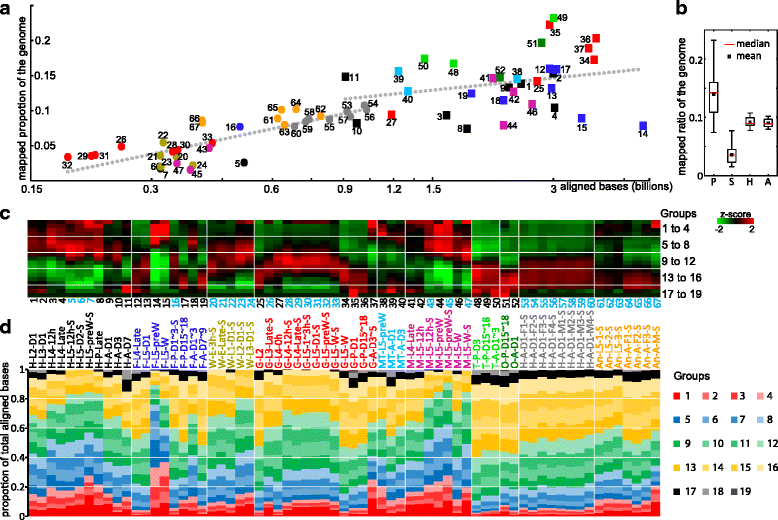
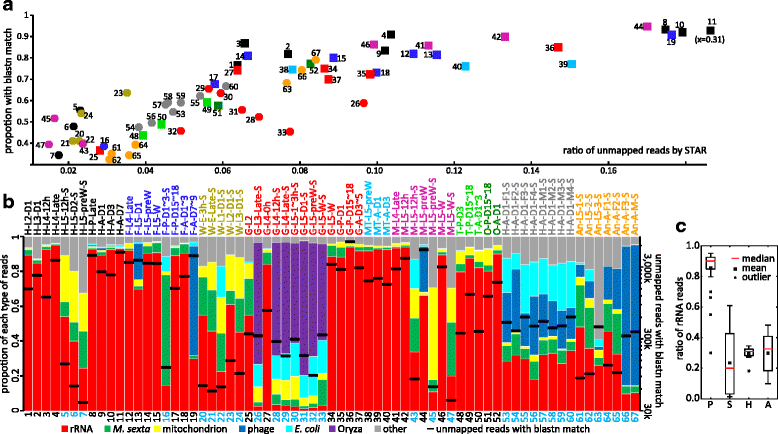
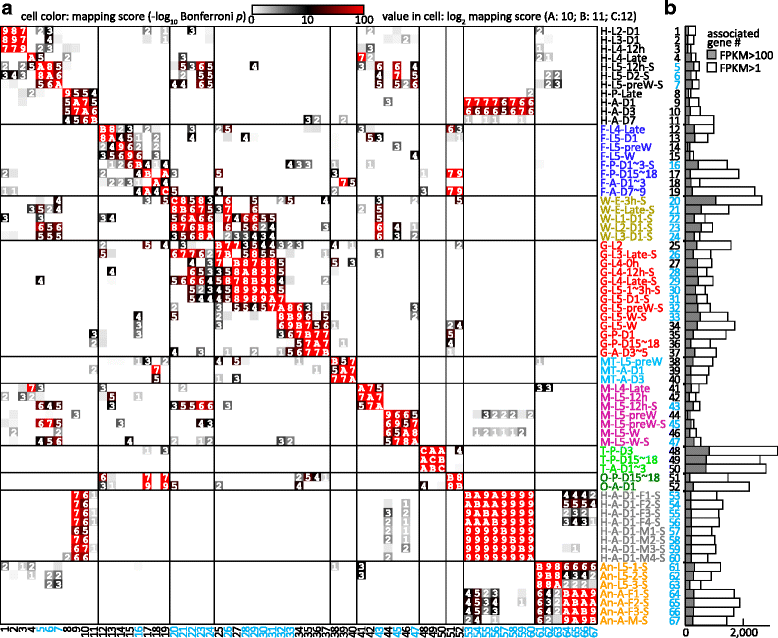
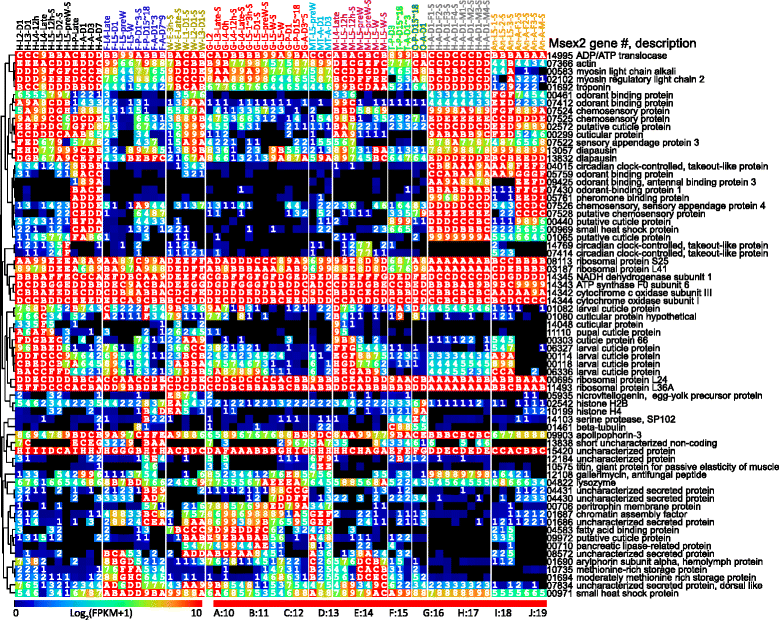
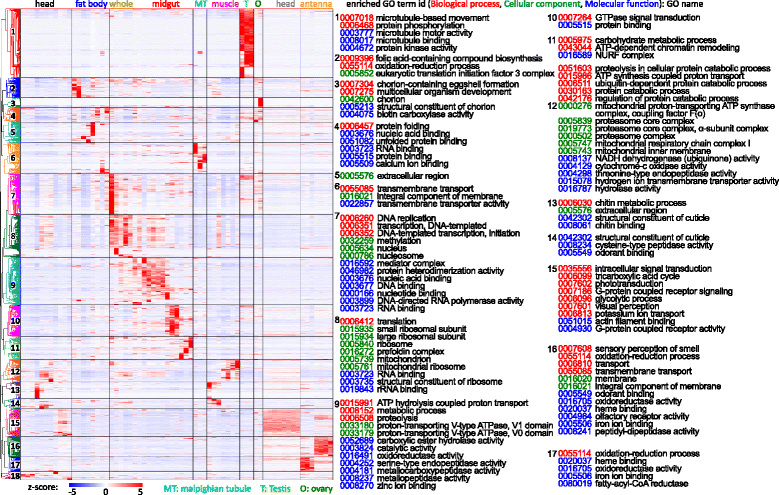
Results: We obtained the transcriptome data, analyzed the raw reads in relation to the assembled genome, and generated heatmaps for clustered genes. Library characteristics (tissues, stages), number of mapped bases, and sequencing methods affected the observed percentages of genome transcription. While up to 40% of the reads were not mapped to the genome in the initial Cufflinks gene modeling, we identified the causes for the mapping failure and reduced the number of non-mappable reads to <8%. Similarities between libraries, measured based on library-correlated genes, clearly identified differences among tissues or life stages. We calculated gene expression levels, analyzed the most abundantly expressed genes in the libraries. Furthermore, we analyzed tissue-specific gene expression and identified 18 groups of genes with distinct expression patterns.
Conclusion: We performed a thorough analysis of the 67 RNA-seq datasets to characterize new genomic features of M. sexta. Integrated knowledge of gene functions and expression features will facilitate future functional studies in this biochemical model insect.
Keywords: Insect genome; Tobacco hornworm; Transcriptome.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
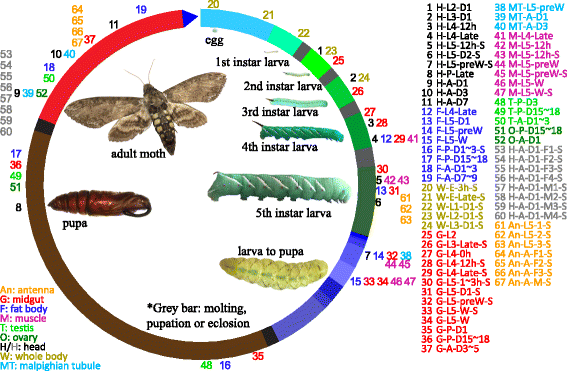
 ), fat body (
), fat body ( ), whole body (
), whole body ( ), midgut (
), midgut ( ), Malpighian tubule (
), Malpighian tubule ( ), muscle (
), muscle ( ), testis (
), testis ( ), ovary (
), ovary ( ), and antenna (
), and antenna ( ). The second part indicates major stages of the insect, i.e. embryo (E), 1st to 5th instar larvae (L1 − L5), pupae (P), and adults (A). In the third part, “D” stands for day, “h” for hour, “preW” for pre-wandering, “W” for wandering, “M” for male, and “F” for female. “S” in the last part of library names indicates single-end sequencing; no “S” in the end indicates paired-end sequencing. The cDNA libraries represent the following tissues and stages: head (H) [1. 2nd (instar) L (larvae), D1 (day 1); 2. 3rd L, D1; 3. 4th L, 12 h (hour); 4. 4th L, late; 5. 5th L, D0.5; 6. 5th L, D2; 7. 5th L, preW (pre-wandering); 8. P (pupae), late; 9. A (adults), D1; 10. A, D3; 11. A, D7], fat body (
). The second part indicates major stages of the insect, i.e. embryo (E), 1st to 5th instar larvae (L1 − L5), pupae (P), and adults (A). In the third part, “D” stands for day, “h” for hour, “preW” for pre-wandering, “W” for wandering, “M” for male, and “F” for female. “S” in the last part of library names indicates single-end sequencing; no “S” in the end indicates paired-end sequencing. The cDNA libraries represent the following tissues and stages: head (H) [1. 2nd (instar) L (larvae), D1 (day 1); 2. 3rd L, D1; 3. 4th L, 12 h (hour); 4. 4th L, late; 5. 5th L, D0.5; 6. 5th L, D2; 7. 5th L, preW (pre-wandering); 8. P (pupae), late; 9. A (adults), D1; 10. A, D3; 11. A, D7], fat body ( ) [12. 4th L, late; 13. 5th L, D1; 14. 5th L, preW; 15. 5th L, W (wandering); 16. P, D1–3; 17. P, D15–18; 18. A, D1–3; 19. A, D7–9], whole body (W) [20. E (embryo), 3 h; 21. E, late; 22. 1st L; 23. 2nd L; 24. 3rd L], midgut (
) [12. 4th L, late; 13. 5th L, D1; 14. 5th L, preW; 15. 5th L, W (wandering); 16. P, D1–3; 17. P, D15–18; 18. A, D1–3; 19. A, D7–9], whole body (W) [20. E (embryo), 3 h; 21. E, late; 22. 1st L; 23. 2nd L; 24. 3rd L], midgut ( ) (25. 2nd L; 26. 3rd L; 27. 4th L, 0 h; 28. 4th L, 12 h; 29. 4th L, late; 30. 5th L, 1–3 h; 31. 5th L, 24 h; 32. 5th L, preW; 33–34. 5th L, W; 35. P, D1; 36. P, D15–18; 37. A, D3–5), Malpighian tubule (
) (25. 2nd L; 26. 3rd L; 27. 4th L, 0 h; 28. 4th L, 12 h; 29. 4th L, late; 30. 5th L, 1–3 h; 31. 5th L, 24 h; 32. 5th L, preW; 33–34. 5th L, W; 35. P, D1; 36. P, D15–18; 37. A, D3–5), Malpighian tubule ( ) (38. 5th L, preW; 39. A, D1; 40. A, D3), muscle (
) (38. 5th L, preW; 39. A, D1; 40. A, D3), muscle ( ) (41. 4th L, late; 42–43. 5th L, 12 h; 44–45. 5th L, preW; 46–47. 5th L, W), testis (
) (41. 4th L, late; 42–43. 5th L, 12 h; 44–45. 5th L, preW; 46–47. 5th L, W), testis ( ) (48. P, D3; 49. P, D15–18; 50. A, D1–3), ovary (
) (48. P, D3; 49. P, D15–18; 50. A, D1–3), ovary ( ) (51. P, D15–18; 52. A, D1), head (
) (51. P, D15–18; 52. A, D1), head ( ) [53–56. A, D1, F (female); 57–60, A, D1, M (male)], antenna (
) [53–56. A, D1, F (female); 57–60, A, D1, M (male)], antenna ( ) (61–63, 5th L; 64–66, A, F; 67, A, M)
) (61–63, 5th L; 64–66, A, F; 67, A, M)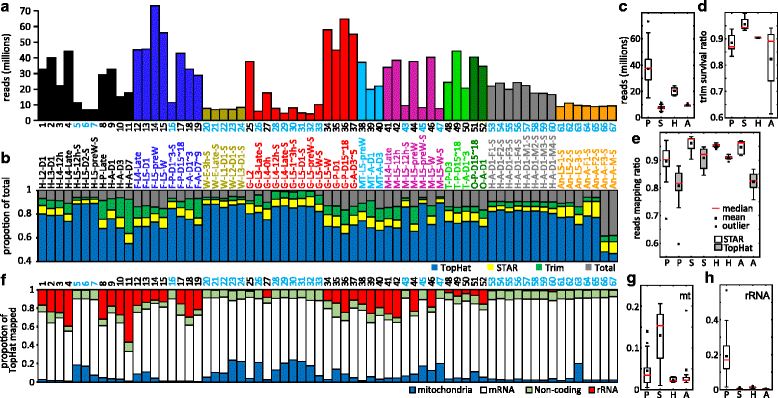
References
-
- Reinecke JP, Buckner J, Grugel S. Life cycle of laboratory-reared tobacco hornworms, Manduca sexta, a study of development and behavior, using time-lapse cinematography. Biol Bull. 1980;158(1):129–140. doi: 10.2307/1540764. - DOI
-
- Tetreau G, Cao X, Chen YR, Muthukrishnan S, Jiang H, Blissard GW, Kanost MR, Wang P. Overview of chitin metabolism enzymes in Manduca sexta: identification, domain organization, phylogenetic analysis and gene expression. Insect Biochem Mol Biol. 2015;62:114–126. doi: 10.1016/j.ibmb.2015.01.006. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

