Imputation of missing genotypes within LD-blocks relying on the basic coalescent and beyond: consideration of population growth and structure
- PMID: 29041903
- PMCID: PMC5646149
- DOI: 10.1186/s12864-017-4208-2
Imputation of missing genotypes within LD-blocks relying on the basic coalescent and beyond: consideration of population growth and structure
Abstract
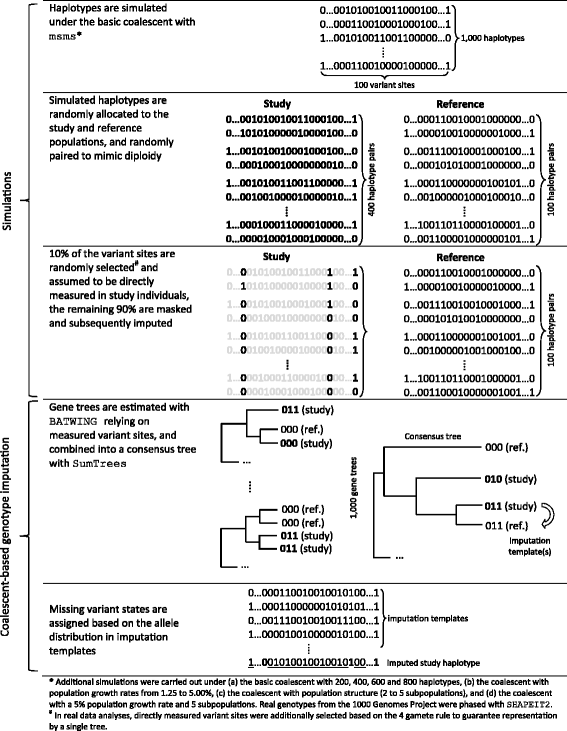
Background: Genotypes not directly measured in genetic studies are often imputed to improve statistical power and to increase mapping resolution. The accuracy of standard imputation techniques strongly depends on the similarity of linkage disequilibrium (LD) patterns in the study and reference populations. Here we develop a novel approach for genotype imputation in low-recombination regions that relies on the coalescent and permits to explicitly account for population demographic factors. To test the new method, study and reference haplotypes were simulated and gene trees were inferred under the basic coalescent and also considering population growth and structure. The reference haplotypes that first coalesced with study haplotypes were used as templates for genotype imputation. Computer simulations were complemented with the analysis of real data. Genotype concordance rates were used to compare the accuracies of coalescent-based and standard (IMPUTE2) imputation.
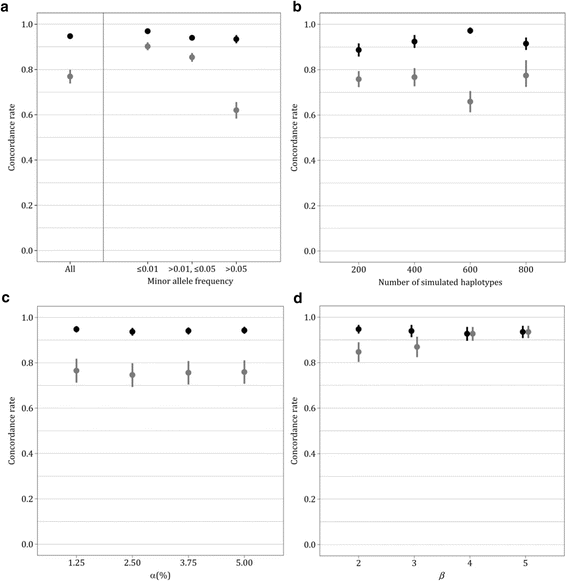
Results: Simulations revealed that, in LD-blocks, imputation accuracy relying on the basic coalescent was higher and less variable than with IMPUTE2. Explicit consideration of population growth and structure, even if present, did not practically improve accuracy. The advantage of coalescent-based over standard imputation increased with the minor allele frequency and it decreased with population stratification. Results based on real data indicated that, even in low-recombination regions, further research is needed to incorporate recombination in coalescence inference, in particular for studies with genetically diverse and admixed individuals.
Conclusions: To exploit the full potential of coalescent-based methods for the imputation of missing genotypes in genetic studies, further methodological research is needed to reduce computer time, to take into account recombination, and to implement these methods in user-friendly computer programs. Here we provide reproducible code which takes advantage of publicly available software to facilitate further developments in the field.
Keywords: Coalescent theory; Genotype imputation; Imputation accuracy; Linkage disequilibrium; Population growth; Population structure.
Conflict of interest statement
Ethics approval and consent to participate
Does not apply since this study has not involved plants, animals or humans directly.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Figures
Similar articles
-
Accuracy of genotype imputation in Nelore cattle.Genet Sel Evol. 2014 Oct 10;46(1):69. doi: 10.1186/s12711-014-0069-1. Genet Sel Evol. 2014. PMID: 25927950 Free PMC article.
-
A generic coalescent-based framework for the selection of a reference panel for imputation.Genet Epidemiol. 2010 Dec;34(8):773-82. doi: 10.1002/gepi.20505. Genet Epidemiol. 2010. PMID: 21058333 Free PMC article.
-
Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation.Am J Hum Genet. 2005 Mar;76(3):449-62. doi: 10.1086/428594. Epub 2005 Jan 31. Am J Hum Genet. 2005. PMID: 15700229 Free PMC article.
-
Two-stage strategy using denoising autoencoders for robust reference-free genotype imputation with missing input genotypes.J Hum Genet. 2024 Oct;69(10):511-518. doi: 10.1038/s10038-024-01261-6. Epub 2024 Jun 25. J Hum Genet. 2024. PMID: 38918526 Free PMC article. Review.
-
On selecting markers for association studies: patterns of linkage disequilibrium between two and three diallelic loci.Genet Epidemiol. 2003 Jan;24(1):57-67. doi: 10.1002/gepi.10217. Genet Epidemiol. 2003. PMID: 12508256 Review.
Cited by
-
Comparing genomic studies in animal breeding and human genetics: focus on disease-related traits in livestock - A review.Anim Biosci. 2025 Feb;38(2):189-197. doi: 10.5713/ab.24.0487. Epub 2024 Oct 24. Anim Biosci. 2025. PMID: 39483033 Free PMC article.
-
A Pipeline for Phasing and Genotype Imputation on Mixed Human Data (Parents-Offspring Trios and Unrelated Subjects) by Reviewing Current Methods and Software.Life (Basel). 2022 Dec 5;12(12):2030. doi: 10.3390/life12122030. Life (Basel). 2022. PMID: 36556394 Free PMC article. Review.
-
Review: Computational analysis of human skeletal remains in ancient DNA and forensic genetics.iScience. 2023 Oct 4;26(11):108066. doi: 10.1016/j.isci.2023.108066. eCollection 2023 Nov 17. iScience. 2023. PMID: 37927550 Free PMC article. Review.
-
Genetic Basis of Low-Salinity Tolerance in the Pacific Oyster (Crassostrea gigas) as Revealed by Estimation of Genetic Parameters and Genome-Wide Association Study.Mar Biotechnol (NY). 2025 May 23;27(3):88. doi: 10.1007/s10126-025-10465-6. Mar Biotechnol (NY). 2025. PMID: 40407927
-
Comparing the effect of imputation reference panel composition in four distinct Latin American cohorts.bioRxiv [Preprint]. 2024 Apr 15:2024.04.11.589057. doi: 10.1101/2024.04.11.589057. bioRxiv. 2024. PMID: 38659746 Free PMC article. Preprint.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

