Analysis of the genome of the New Zealand giant collembolan (Holacanthella duospinosa) sheds light on hexapod evolution
- PMID: 29041914
- PMCID: PMC5644144
- DOI: 10.1186/s12864-017-4197-1
Analysis of the genome of the New Zealand giant collembolan (Holacanthella duospinosa) sheds light on hexapod evolution
Abstract
Background: The New Zealand collembolan genus Holacanthella contains the largest species of springtails (Collembola) in the world. Using Illumina technology we have sequenced and assembled a draft genome and transcriptome from Holacanthella duospinosa (Salmon). We have used this annotated assembly to investigate the genetic basis of a range of traits critical to the evolution of the Hexapoda, the phylogenetic position of H. duospinosa and potential horizontal gene transfer events.
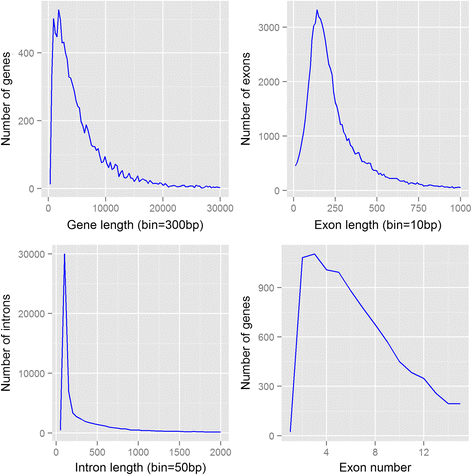
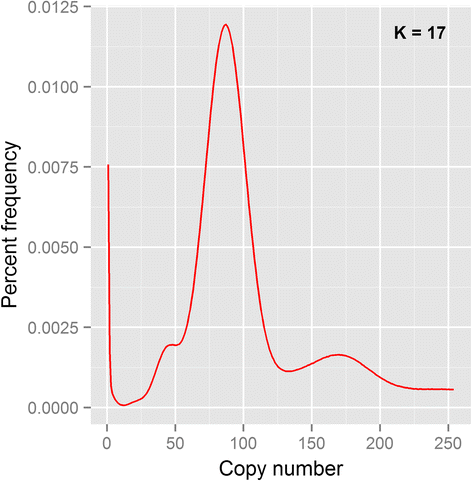
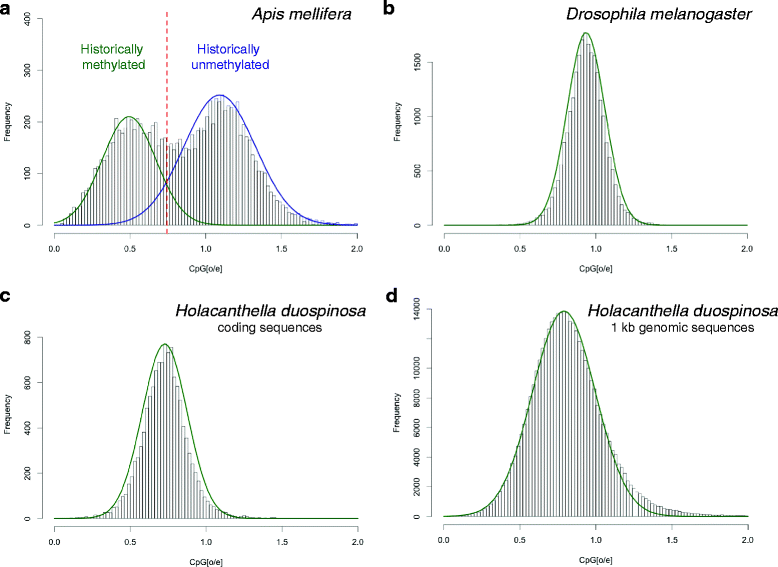
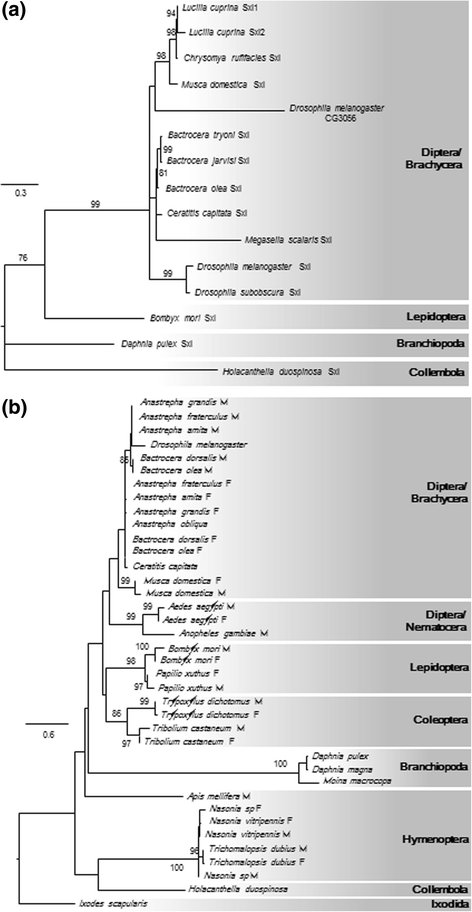
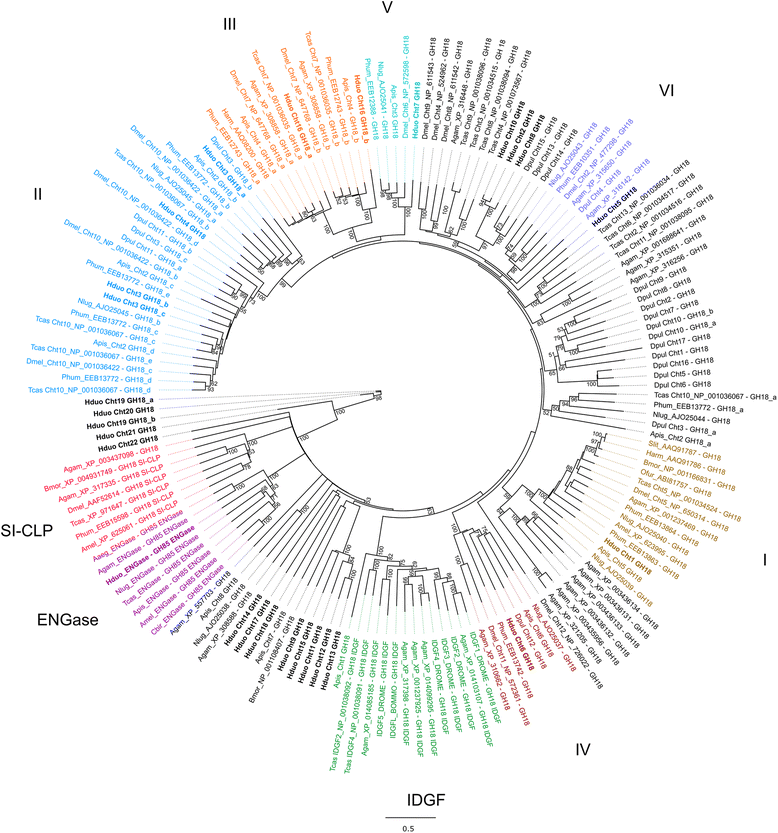
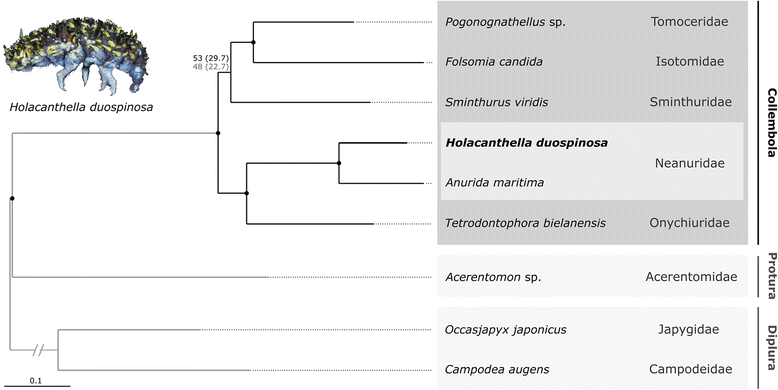
Results: Our genome assembly was ~375 Mbp in size with a scaffold N50 of ~230 Kbp and sequencing coverage of ~180×. DNA elements, LTRs and simple repeats and LINEs formed the largest components and SINEs were very rare. Phylogenomics (370,877 amino acids) placed H. duospinosa within the Neanuridae. We recovered orthologs of the conserved sex determination genes thought to play a role in sex determination. Analysis of CpG content suggested the absence of DNA methylation, and consistent with this we were unable to detect orthologs of the DNA methyltransferase enzymes. The small subunit rRNA gene contained a possible retrotransposon. The Hox gene complex was broken over two scaffolds. For chemosensory ability, at least 15 and 18 ionotropic glutamate and gustatory receptors were identified, respectively. However, we were unable to identify any odorant receptors or their obligate co-receptor Orco. Twenty-three chitinase-like genes were identified from the assembly. Members of this multigene family may play roles in the digestion of fungal cell walls, a common food source for these saproxylic organisms. We also detected 59 and 96 genes that blasted to bacteria and fungi, respectively, but were located on scaffolds that otherwise contained arthropod genes.
Conclusions: The genome of H. duospinosa contains some unusual features including a Hox complex broken over two scaffolds, in a different manner to other arthropod species, a lack of odorant receptor genes and an apparent lack of environmentally responsive DNA methylation, unlike many other arthropods. Our detection of candidate horizontal gene transfer candidates confirms that this phenomenon is occurring across Collembola. These findings allow us to narrow down the regions of the arthropod phylogeny where key innovations have occurred that have facilitated the evolutionary success of Hexapoda.
Keywords: Chemoreceptors; Developmental biology; Epigenetics; Genome assembly; Hexapoda; Horizontal gene transfer; Methylation; Neanuridae; Phylogenomics; RNA; Sex determination.
Conflict of interest statement
Ethics approval and consent to participate
Approval for the research was given by Ngāti Manuhiri, who are kaitiaki (guardians) of Te Hauturu-o-Toi, from where the specimens were collected. Specimens were collected under a permit issued by the Department of Conservation (AK-29386-FAU).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
A High-quality Draft Genome Assembly of Sinella curviseta: A Soil Model Organism (Collembola).Genome Biol Evol. 2019 Feb 1;11(2):521-530. doi: 10.1093/gbe/evz013. Genome Biol Evol. 2019. PMID: 30668671 Free PMC article.
-
Coping with living in the soil: the genome of the parthenogenetic springtail Folsomia candida.BMC Genomics. 2017 Jun 28;18(1):493. doi: 10.1186/s12864-017-3852-x. BMC Genomics. 2017. PMID: 28659179 Free PMC article.
-
Chromosomal-Level Genome Assembly of the Springtail Tomocerus qinae (Collembola: Tomoceridae).Genome Biol Evol. 2022 Apr 10;14(4):evac039. doi: 10.1093/gbe/evac039. Genome Biol Evol. 2022. PMID: 35298623 Free PMC article.
-
Molecular Evolution of the Major Arthropod Chemoreceptor Gene Families.Annu Rev Entomol. 2019 Jan 7;64:227-242. doi: 10.1146/annurev-ento-020117-043322. Epub 2018 Oct 12. Annu Rev Entomol. 2019. PMID: 30312552 Review.
-
Advances in genome sequencing reveal changes in gene content that contribute to arthropod macroevolution.Dev Genes Evol. 2023 Dec;233(2):59-76. doi: 10.1007/s00427-023-00712-y. Epub 2023 Nov 20. Dev Genes Evol. 2023. PMID: 37982820 Review.
Cited by
-
Disparate expression specificities coded by a shared Hox-C enhancer.Elife. 2020 Apr 28;9:e39876. doi: 10.7554/eLife.39876. Elife. 2020. PMID: 32342858 Free PMC article.
-
Diversity of the Antimicrobial Peptide Genes in Collembola.Insects. 2023 Feb 21;14(3):215. doi: 10.3390/insects14030215. Insects. 2023. PMID: 36975900 Free PMC article.
-
Genome expansion of an obligate parthenogenesis-associated Wolbachia poses an exception to the symbiont reduction model.BMC Genomics. 2019 Feb 6;20(1):106. doi: 10.1186/s12864-019-5492-9. BMC Genomics. 2019. PMID: 30727958 Free PMC article.
-
Evolutionary emergence of Hairless as a novel component of the Notch signaling pathway.Elife. 2019 Sep 23;8:e48115. doi: 10.7554/eLife.48115. Elife. 2019. PMID: 31545167 Free PMC article.
-
A High-quality Draft Genome Assembly of Sinella curviseta: A Soil Model Organism (Collembola).Genome Biol Evol. 2019 Feb 1;11(2):521-530. doi: 10.1093/gbe/evz013. Genome Biol Evol. 2019. PMID: 30668671 Free PMC article.
References
-
- D'Haese CA. Morphological appraisal of Collembola phylogeny with special emphasis on Poduromorpha and a test of the aquatic origin hypothesis. Zool Scr. 2003;32(6):563–586.
-
- Misof B, Liu SL, Meusemann K, Peters RS, Donath A, Mayer C, Frandsen PB, Ware J, Flouri T, Beutel RG, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346(6210):763–767. - PubMed
-
- Stevens MI, Greenslade P, Hogg ID, Sunnucks P. Southern hemisphere springtails: could any have survived glaciation of Antarctica? Mol Biol Evol. 2006;23(5):874–882. - PubMed
-
- Nardi F, Spinsanti G, Boore JL, Carapelli A, Dallai R, Frati F. Response to comment on "hexapod origins: monophyletic or paraphyletic? ". Science. 2003;301(5639):1482. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

