Differential expression of porcine microRNAs in African swine fever virus infected pigs: a proof-of-concept study
- PMID: 29041944
- PMCID: PMC5646143
- DOI: 10.1186/s12985-017-0864-8
Differential expression of porcine microRNAs in African swine fever virus infected pigs: a proof-of-concept study
Abstract
Background: African swine fever (ASF) is a re-expanding devastating viral disease currently threatening the pig industry worldwide. MicroRNAs are a class of 17-25 nucleotide non- coding RNAs that have been shown to have critical functions in a wide variety of biological processes, such as cell differentiation, cell cycle regulation, carcinogenesis, apoptosis, regulation of immunity as well as in viral infections by cleavage or translational repression of mRNAs. Nevertheless, there is no information about miRNA expression in an ASFV infection.
Methods: In this proof-of-concept study, we have analyzed miRNAs expressed in spleen and submandibular lymph node of experimentally infected pigs with a virulent (E75) or its derived attenuated (E75CV1) ASFV strain, as well as, at different times post-infection with the virulent strain, by high throughput sequencing of small RNA libraries.
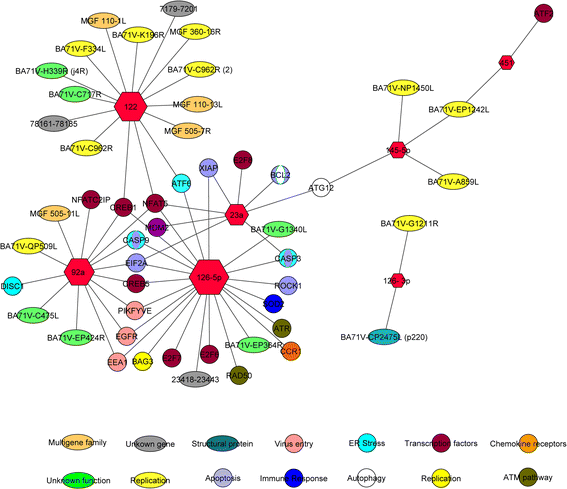
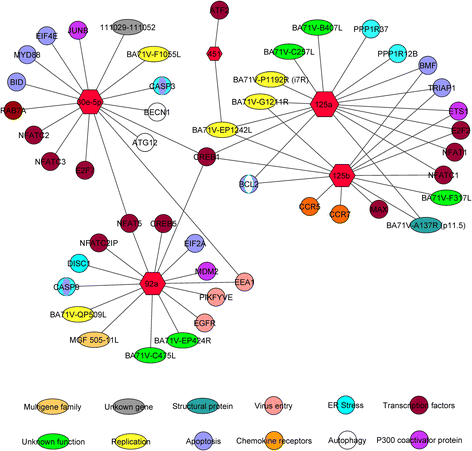
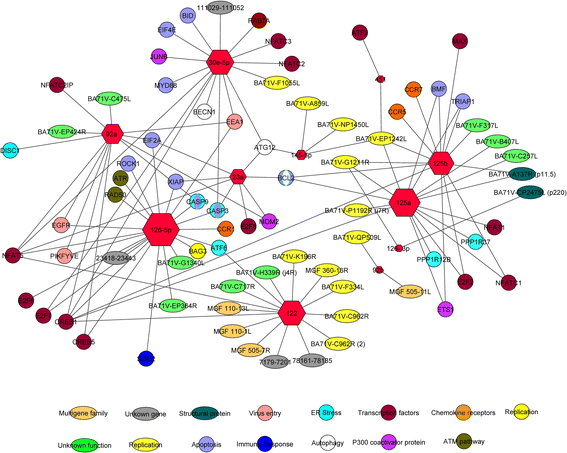
Results: Spleen presented a more differential expression pattern than lymph nodes in an ASFV infection. Of the most abundant miRNAs, 12 were differentially expressed in both tissues at two different times in infected animals with the virulent strain. Of these, miR-451, miR-145-5p, miR-181a and miR-122 presented up-regulation at late times post-infection while miR-92a, miR-23a, miR-92b-3p, miR-126-5p, miR-126-3p, miR-30d, miR-23b and miR-92c showed down-regulation. Of the 8 differentially expressed miRNAs identified at the same time post-infection in infected animals with the virulent strain compared with animals infected with its attenuated strain, miR-126-5p, miR-92c, miR-92a, miR-30e-5p and miR-500a-5p presented up-regulation whereas miR-125b, miR-451 and miR-125a were down-regulated. All these miRNAs have been shown to be associated with cellular genes involved in pathways related to the immune response, virus-host interactions as well as with several viral genes.
Conclusion: The study of miRNA expression will contribute to a better understanding of African swine fever virus pathogenesis, essential in the development of any disease control strategy.
Conflict of interest statement
Ethics approval and consent to participate
All procedures were carried out in strict accordance with the guidelines of the institutional animal ethics committee of Universitat Autònoma de Barcelona (UAB), with the permit number DMAH-5796 for this specific study, according to the Spanish and European animal experimentation ethics law. Animals were euthanized following the European Directive 2010/63/EU.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Identification of a Functional Small Noncoding RNA of African Swine Fever Virus.J Virol. 2020 Oct 14;94(21):e01515-20. doi: 10.1128/JVI.01515-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32796064 Free PMC article.
-
African swine fever virus does not express viral microRNAs in experimentally infected pigs.BMC Vet Res. 2018 Sep 3;14(1):268. doi: 10.1186/s12917-018-1601-2. BMC Vet Res. 2018. PMID: 30176871 Free PMC article.
-
Small RNA sequencing and profiling of serum-derived exosomes from African swine fever virus-infected pigs.J Anim Sci. 2023 Jan 3;101:skac400. doi: 10.1093/jas/skac400. J Anim Sci. 2023. PMID: 36478238 Free PMC article.
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
-
[African swine fever in Russian Federation].Vopr Virusol. 2012 Sep-Oct;57(5):4-10. Vopr Virusol. 2012. PMID: 23248852 Review. Russian.
Cited by
-
The transcriptomic insight into the differential susceptibility of African Swine Fever in inbred pigs.Sci Rep. 2024 Mar 11;14(1):5944. doi: 10.1038/s41598-024-56569-2. Sci Rep. 2024. PMID: 38467747 Free PMC article.
-
MicroRNAs as Biomarkers for Animal Health and Welfare in Livestock.Front Vet Sci. 2020 Dec 18;7:578193. doi: 10.3389/fvets.2020.578193. eCollection 2020. Front Vet Sci. 2020. PMID: 33392281 Free PMC article. Review.
-
Identifying miRNA-mRNA regulatory networks on extreme n-6/n-3 polyunsaturated fatty acid ratio expression profiles in porcine skeletal muscle.PLoS One. 2023 May 4;18(5):e0283231. doi: 10.1371/journal.pone.0283231. eCollection 2023. PLoS One. 2023. PMID: 37141193 Free PMC article.
-
Identification of a Functional Small Noncoding RNA of African Swine Fever Virus.J Virol. 2020 Oct 14;94(21):e01515-20. doi: 10.1128/JVI.01515-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32796064 Free PMC article.
-
African swine fever virus does not express viral microRNAs in experimentally infected pigs.BMC Vet Res. 2018 Sep 3;14(1):268. doi: 10.1186/s12917-018-1601-2. BMC Vet Res. 2018. PMID: 30176871 Free PMC article.
References
-
- Eustace Montgomery R. On a form of swine fever occurring in British East Africa (Kenya Colony) J Comp Pathol Ther. 1921;34:159–191. doi: 10.1016/S0368-1742(21)80031-4. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

