Enrichment allows identification of diverse, rare elements in metagenomic resistome-virulome sequencing
- PMID: 29041965
- PMCID: PMC5645900
- DOI: 10.1186/s40168-017-0361-8
Enrichment allows identification of diverse, rare elements in metagenomic resistome-virulome sequencing
Abstract
Background: Shotgun metagenomic sequencing is increasingly utilized as a tool to evaluate ecological-level dynamics of antimicrobial resistance and virulence, in conjunction with microbiome analysis. Interest in use of this method for environmental surveillance of antimicrobial resistance and pathogenic microorganisms is also increasing. In published metagenomic datasets, the total of all resistance- and virulence-related sequences accounts for < 1% of all sequenced DNA, leading to limitations in detection of low-abundance resistome-virulome elements. This study describes the extent and composition of the low-abundance portion of the resistome-virulome, using a bait-capture and enrichment system that incorporates unique molecular indices to count DNA molecules and correct for enrichment bias.
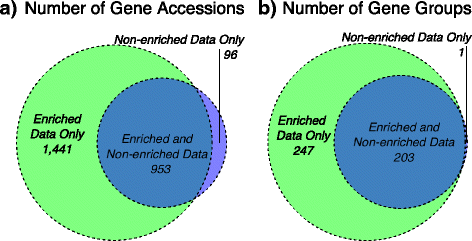
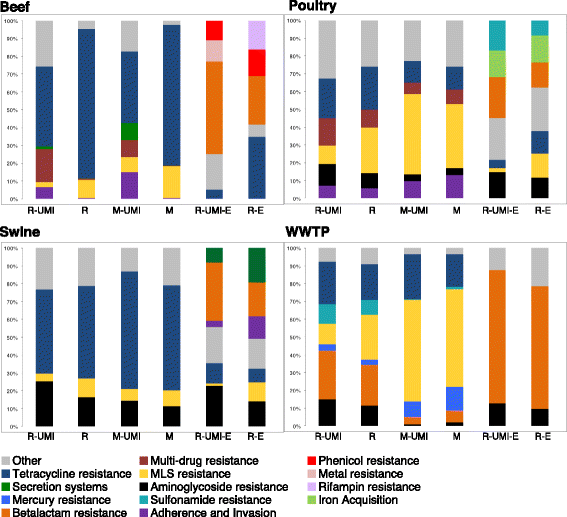
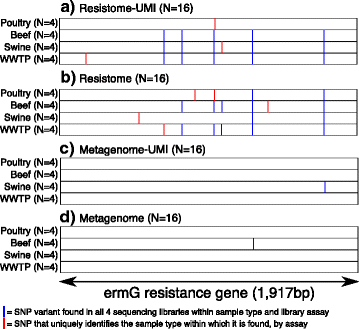
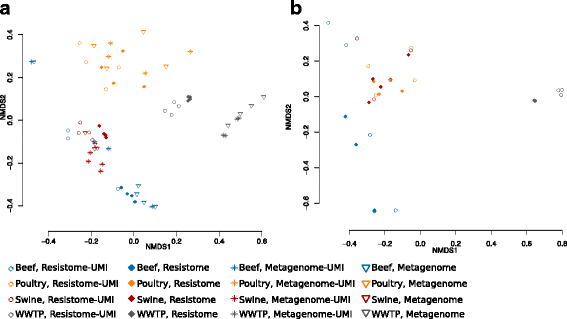
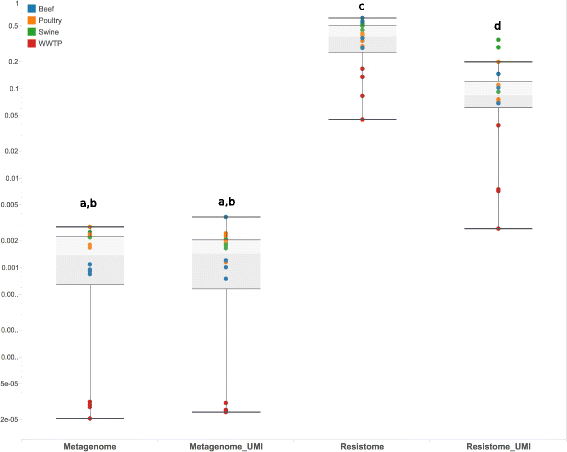
Results: The use of the bait-capture and enrichment system significantly increased on-target sequencing of the resistome-virulome, enabling detection of an additional 1441 gene accessions and revealing a low-abundance portion of the resistome-virulome that was more diverse and compositionally different than that detected by more traditional metagenomic assays. The low-abundance portion of the resistome-virulome also contained resistance genes with public health importance, such as extended-spectrum betalactamases, that were not detected using traditional shotgun metagenomic sequencing. In addition, the use of the bait-capture and enrichment system enabled identification of rare resistance gene haplotypes that were used to discriminate between sample origins.
Conclusions: These results demonstrate that the rare resistome-virulome contains valuable and unique information that can be utilized for both surveillance and population genetic investigations of resistance. Access to the rare resistome-virulome using the bait-capture and enrichment system validated in this study can greatly advance our understanding of microbiome-resistome dynamics.
Keywords: Antimicrobial resistance; Microbial ecology; Molecular enrichment; Rare microbiome; Resistome.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
Multiplexed Target Enrichment Enables Efficient and In-Depth Analysis of Antimicrobial Resistome in Metagenomes.Microbiol Spectr. 2022 Dec 21;10(6):e0229722. doi: 10.1128/spectrum.02297-22. Epub 2022 Oct 26. Microbiol Spectr. 2022. PMID: 36287061 Free PMC article.
-
Exploiting a targeted resistome sequencing approach in assessing antimicrobial resistance in retail foods.Environ Microbiome. 2023 Mar 29;18(1):25. doi: 10.1186/s40793-023-00482-0. Environ Microbiome. 2023. PMID: 36991496 Free PMC article.
-
Capturing the Resistome: a Targeted Capture Method To Reveal Antibiotic Resistance Determinants in Metagenomes.Antimicrob Agents Chemother. 2019 Dec 20;64(1):e01324-19. doi: 10.1128/AAC.01324-19. Print 2019 Dec 20. Antimicrob Agents Chemother. 2019. PMID: 31611361 Free PMC article.
-
Translational metagenomics and the human resistome: confronting the menace of the new millennium.J Mol Med (Berl). 2017 Jan;95(1):41-51. doi: 10.1007/s00109-016-1478-0. Epub 2016 Oct 20. J Mol Med (Berl). 2017. PMID: 27766372 Free PMC article. Review.
-
Metagenomic Insights into Transferable Antibiotic Resistance in Oral Bacteria.J Dent Res. 2016 Aug;95(9):969-76. doi: 10.1177/0022034516648944. Epub 2016 May 16. J Dent Res. 2016. PMID: 27183895 Review.
Cited by
-
A unified catalog of 204,938 reference genomes from the human gut microbiome.Nat Biotechnol. 2021 Jan;39(1):105-114. doi: 10.1038/s41587-020-0603-3. Epub 2020 Jul 20. Nat Biotechnol. 2021. PMID: 32690973 Free PMC article.
-
Multiplexed Target Enrichment Enables Efficient and In-Depth Analysis of Antimicrobial Resistome in Metagenomes.Microbiol Spectr. 2022 Dec 21;10(6):e0229722. doi: 10.1128/spectrum.02297-22. Epub 2022 Oct 26. Microbiol Spectr. 2022. PMID: 36287061 Free PMC article.
-
Exploiting a targeted resistome sequencing approach in assessing antimicrobial resistance in retail foods.Environ Microbiome. 2023 Mar 29;18(1):25. doi: 10.1186/s40793-023-00482-0. Environ Microbiome. 2023. PMID: 36991496 Free PMC article.
-
Modeling the limits of detection for antimicrobial resistance genes in agri-food samples: a comparative analysis of bioinformatics tools.BMC Microbiol. 2024 Jan 20;24(1):31. doi: 10.1186/s12866-023-03148-6. BMC Microbiol. 2024. PMID: 38245666 Free PMC article.
-
The impact of sequencing depth on the inferred taxonomic composition and AMR gene content of metagenomic samples.Environ Microbiome. 2019 Oct 24;14(1):7. doi: 10.1186/s40793-019-0347-1. Environ Microbiome. 2019. PMID: 33902704 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

