Plastic Transcriptomes Stabilize Immunity to Pathogen Diversity: The Jasmonic Acid and Salicylic Acid Networks within the Arabidopsis/ Botrytis Pathosystem
- PMID: 29042403
- PMCID: PMC5728128
- DOI: 10.1105/tpc.17.00348
Plastic Transcriptomes Stabilize Immunity to Pathogen Diversity: The Jasmonic Acid and Salicylic Acid Networks within the Arabidopsis/ Botrytis Pathosystem
Abstract
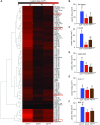
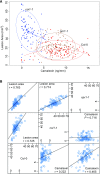
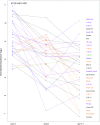
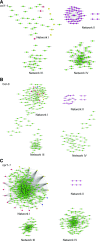
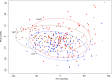
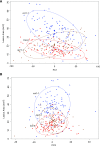
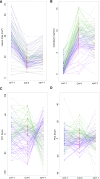
To respond to pathogen attack, selection and associated evolution has led to the creation of plant immune system that are a highly effective and inducible defense system. Central to this system are the plant defense hormones jasmonic acid (JA) and salicylic acid (SA) and crosstalk between the two, which may play an important role in defense responses to specific pathogens or even genotypes. Here, we used the Arabidopsis thaliana-Botrytis cinerea pathosystem to test how the host's defense system functions against genetic variation in a pathogen. We measured defense-related phenotypes and transcriptomic responses in Arabidopsis wild-type Col-0 and JA- and SA-signaling mutants, coi1-1 and npr1-1, individually challenged with 96 diverse B. cinerea isolates. Those data showed genetic variation in the pathogen influences on all components within the plant defense system at the transcriptional level. We identified four gene coexpression networks and two vectors of defense variation triggered by genetic variation in B. cinerea This showed that the JA and SA signaling pathways functioned to constrain/canalize the range of virulence in the pathogen population, but the underlying transcriptomic response was highly plastic. These data showed that plants utilize major defense hormone pathways to buffer disease resistance, but not the metabolic or transcriptional responses to genetic variation within a pathogen.
© 2017 American Society of Plant Biologists. All rights reserved.
Figures


Similar articles
-
Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant.Plant Cell. 2003 Dec;15(12):2952-65. doi: 10.1105/tpc.017301. Epub 2003 Nov 13. Plant Cell. 2003. PMID: 14615603 Free PMC article.
-
Botrytis cinerea B05.10 promotes disease development in Arabidopsis by suppressing WRKY33-mediated host immunity.Plant Cell Environ. 2017 Oct;40(10):2189-2206. doi: 10.1111/pce.13022. Epub 2017 Aug 17. Plant Cell Environ. 2017. PMID: 28708934
-
Ethylene and jasmonic acid signaling affect the NPR1-independent expression of defense genes without impacting resistance to Pseudomonas syringae and Peronospora parasitica in the Arabidopsis ssi1 mutant.Mol Plant Microbe Interact. 2003 Jul;16(7):588-99. doi: 10.1094/MPMI.2003.16.7.588. Mol Plant Microbe Interact. 2003. PMID: 12848424
-
Signaling Crosstalk between Salicylic Acid and Ethylene/Jasmonate in Plant Defense: Do We Understand What They Are Whispering?Int J Mol Sci. 2019 Feb 4;20(3):671. doi: 10.3390/ijms20030671. Int J Mol Sci. 2019. PMID: 30720746 Free PMC article. Review.
-
NPR1 in JazzSet with Pathogen Effectors.Trends Plant Sci. 2018 Jun;23(6):469-472. doi: 10.1016/j.tplants.2018.04.007. Epub 2018 May 9. Trends Plant Sci. 2018. PMID: 29753632 Review.
Cited by
-
A Novel ERF Transcription Factor, ZmERF105, Positively Regulates Maize Resistance to Exserohilum turcicum.Front Plant Sci. 2020 Jun 16;11:850. doi: 10.3389/fpls.2020.00850. eCollection 2020. Front Plant Sci. 2020. PMID: 32612628 Free PMC article.
-
Expression of disease resistance in genetically modified grapevines correlates with the contents of viral sequences in the T-DNA and global genome methylation.Transgenic Res. 2018 Aug;27(4):379-396. doi: 10.1007/s11248-018-0082-1. Epub 2018 Jun 6. Transgenic Res. 2018. PMID: 29876789
-
Bacillus circulans GN03 Alters the Microbiota, Promotes Cotton Seedling Growth and Disease Resistance, and Increases the Expression of Phytohormone Synthesis and Disease Resistance-Related Genes.Front Plant Sci. 2021 Apr 14;12:644597. doi: 10.3389/fpls.2021.644597. eCollection 2021. Front Plant Sci. 2021. PMID: 33936131 Free PMC article.
-
Melatonin enhances cotton immunity to Verticillium wilt via manipulating lignin and gossypol biosynthesis.Plant J. 2019 Nov;100(4):784-800. doi: 10.1111/tpj.14477. Epub 2019 Aug 31. Plant J. 2019. PMID: 31349367 Free PMC article.
-
97 Shades of Gray: Genetic Interactions of the Gray Mold, Botrytis cinerea, with Wild and Domesticated Tomato.Plant Cell. 2019 Feb;31(2):280-281. doi: 10.1105/tpc.19.00030. Epub 2019 Jan 16. Plant Cell. 2019. PMID: 30651347 Free PMC article. No abstract available.
References
-
- An B., Li B., Qin G., Tian S. (2015). Function of small GTPase Rho3 in regulating growth, conidiation and virulence of Botrytis cinerea Fungal Genet. Biol. 75: 46–55. - PubMed
-
- Bednarek P., Piślewska-Bednarek M., Ver Loren van Themaat E., Maddula R.K., Svatoš A., Schulze-Lefert P. (2011). Conservation and clade-specific diversification of pathogen-inducible tryptophan and indole glucosinolate metabolism in Arabidopsis thaliana relatives. New Phytol. 192: 713–726. - PubMed
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. A Stat. Soc. 57: 289–300.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

