Sterol methyltransferase a target for anti-amoeba therapy: towards transition state analog and suicide substrate drug design
- PMID: 29042405
- PMCID: PMC5711494
- DOI: 10.1194/jlr.M079418
Sterol methyltransferase a target for anti-amoeba therapy: towards transition state analog and suicide substrate drug design
Abstract
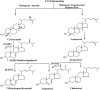
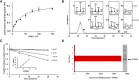
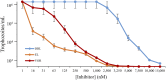
Ergosterol biosynthesis pathways essential to pathogenic protozoa growth and absent from the human host offer new chokepoint targets. Here, we present characterization and cell-based interference of Acanthamoeba spp sterol 24-/28-methylases (SMTs) that catalyze the committed step in C28- and C29-sterol synthesis. Intriguingly, our kinetic analyses suggest that 24-SMT prefers plant cycloartenol whereas 28-SMT prefers 24(28)-methylene lophenol in similar fashion to the substrate preferences of land plant SMT1 and SMT2. Transition state analog-24(R,S),25-epiminolanosterol (EL) and suicide substrate 26,27-dehydrolanosterol (DHL) differentially inhibited trophozoite growth with IC50 values of 7 nM and 6 µM, respectively, and EL yielded 20-fold higher activity than reference drug voriconazole. Against either SMT assayed with native substrate, EL exhibited tight binding ∼Ki 9 nM. Alternatively, DHL is methylated at C26 by 24-SMT that thereby, generates intermediates that complex and inactivate the enzyme, whereas DHL is not productively bound to 28-SMT. Steroidal inhibitors had no effect on human epithelial kidney cell growth or cholesterol biosynthesis at minimum amoebicidal concentrations. We hypothesize the selective inhibition of Acanthamoeba by steroidal inhibitors representing distinct chemotypes may be an efficient strategy for the development of promising compounds to combat amoeba diseases.
Keywords: Acanthamoeba; anti-amoeba drugs; ergosterol biosynthesis inhibitors; phytosterol biosynthesis; sterol C24-methyltransferase.
Copyright © 2017 by the American Society for Biochemistry and Molecular Biology, Inc.
Figures








Similar articles
-
Two families of sterol methyltransferases are involved in the first and the second methylation steps of plant sterol biosynthesis.Eur J Biochem. 1998 Aug 15;256(1):88-96. doi: 10.1046/j.1432-1327.1998.2560088.x. Eur J Biochem. 1998. PMID: 9746350
-
Cloning, mechanistic and functional analysis of a fungal sterol C24-methyltransferase implicated in brassicasterol biosynthesis.Biochim Biophys Acta. 2010 Oct;1801(10):1163-74. doi: 10.1016/j.bbalip.2010.06.007. Epub 2010 Jul 17. Biochim Biophys Acta. 2010. PMID: 20624480
-
Cloning, functional expression and phylogenetic analysis of plant sterol 24C-methyltransferases involved in sitosterol biosynthesis.Phytochemistry. 2009 Dec;70(17-18):1982-98. doi: 10.1016/j.phytochem.2009.09.003. Epub 2009 Oct 8. Phytochemistry. 2009. PMID: 19818974
-
Sterol methyl transferase: enzymology and inhibition.Biochim Biophys Acta. 2000 Dec 15;1529(1-3):63-88. doi: 10.1016/s1388-1981(00)00138-4. Biochim Biophys Acta. 2000. PMID: 11111078 Review.
-
Steroidal triterpenes: design of substrate-based inhibitors of ergosterol and sitosterol synthesis.Molecules. 2009 Nov 18;14(11):4690-706. doi: 10.3390/molecules14114690. Molecules. 2009. PMID: 19924096 Free PMC article. Review.
Cited by
-
Homology Modeling, de Novo Design of Ligands, and Molecular Docking Identify Potential Inhibitors of Leishmania donovani 24-Sterol Methyltransferase.Front Cell Infect Microbiol. 2022 Jun 2;12:859981. doi: 10.3389/fcimb.2022.859981. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35719359 Free PMC article.
-
Druggable Sterol Metabolizing Enzymes in Infectious Diseases: Cell Targets to Therapeutic Leads.Biomolecules. 2024 Feb 20;14(3):249. doi: 10.3390/biom14030249. Biomolecules. 2024. PMID: 38540670 Free PMC article. Review.
-
Identification of Potent and Selective Inhibitors of Acanthamoeba: Structural Insights into Sterol 14α-Demethylase as a Key Drug Target.J Med Chem. 2024 May 9;67(9):7443-7457. doi: 10.1021/acs.jmedchem.4c00303. Epub 2024 Apr 29. J Med Chem. 2024. PMID: 38683753 Free PMC article.
-
Synthesis and Biological Activity of Sterol 14α-Demethylase and Sterol C24-Methyltransferase Inhibitors.Molecules. 2018 Jul 17;23(7):1753. doi: 10.3390/molecules23071753. Molecules. 2018. PMID: 30018257 Free PMC article. Review.
-
Discovery of repurposing drug candidates for the treatment of diseases caused by pathogenic free-living amoebae.PLoS Negl Trop Dis. 2020 Sep 24;14(9):e0008353. doi: 10.1371/journal.pntd.0008353. eCollection 2020 Sep. PLoS Negl Trop Dis. 2020. PMID: 32970675 Free PMC article.
References
-
- Visvesvara G. S. 2010. Free-living amebae as opportunistic agents of human disease. J. Neuroparasitology. 1: 1–13.
-
- Dart J. K., Saw V. P., and Kilvington S.. 2009. Acanthamoeba keratitis: Diagnosis and treatment update 2009. Am. J. Ophthalmol. 148: 487–499.e2. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

