Autophagy induction under carbon starvation conditions is negatively regulated by carbon catabolite repression
- PMID: 29042435
- PMCID: PMC5712628
- DOI: 10.1074/jbc.M117.817510
Autophagy induction under carbon starvation conditions is negatively regulated by carbon catabolite repression
Abstract
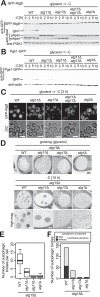
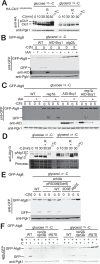
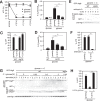
Autophagy is a conserved process in which cytoplasmic components are sequestered for degradation in the vacuole/lysosomes in eukaryotic cells. Autophagy is induced under a variety of starvation conditions, such as the depletion of nitrogen, carbon, phosphorus, zinc, and others. However, apart from nitrogen starvation, it remains unclear how these stimuli induce autophagy. In yeast, for example, it remains contentious whether autophagy is induced under carbon starvation conditions, with reports variously suggesting both induction and lack of induction upon depletion of carbon. We therefore undertook an analysis to account for these inconsistencies, concluding that autophagy is induced in response to abrupt carbon starvation when cells are grown with glycerol but not glucose as the carbon source. We found that autophagy under these conditions is mediated by nonselective degradation that is highly dependent on the autophagosome-associated scaffold proteins Atg11 and Atg17. We also found that the extent of carbon starvation-induced autophagy is positively correlated with cells' oxygen consumption rate, drawing a link between autophagy induction and respiratory metabolism. Further biochemical analyses indicated that maintenance of intracellular ATP levels is also required for carbon starvation-induced autophagy and that autophagy plays an important role in cell viability during prolonged carbon starvation. Our findings suggest that carbon starvation-induced autophagy is negatively regulated by carbon catabolite repression.
Keywords: ATP; autophagy; energy metabolism; glucose; yeast.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


Similar articles
-
Induction of autophagy by phosphate starvation in an Atg11-dependent manner in Saccharomyces cerevisiae.Biochem Biophys Res Commun. 2017 Jan 29;483(1):522-527. doi: 10.1016/j.bbrc.2016.12.112. Epub 2016 Dec 21. Biochem Biophys Res Commun. 2017. PMID: 28013049
-
Atg11 is required for initiation of glucose starvation-induced autophagy.Autophagy. 2020 Dec;16(12):2206-2218. doi: 10.1080/15548627.2020.1719724. Epub 2020 Feb 3. Autophagy. 2020. PMID: 31971848 Free PMC article.
-
Mechanistic Insights into the Role of Atg11 in Selective Autophagy.J Mol Biol. 2020 Jan 3;432(1):104-122. doi: 10.1016/j.jmb.2019.06.017. Epub 2019 Jun 22. J Mol Biol. 2020. PMID: 31238043 Free PMC article. Review.
-
Ecm33 is a novel factor involved in efficient glucose uptake for nutrition-responsive TORC1 signaling in yeast.FEBS Lett. 2017 Nov;591(22):3721-3729. doi: 10.1002/1873-3468.12882. Epub 2017 Oct 27. FEBS Lett. 2017. PMID: 29029364
-
Glucose repression in Saccharomyces cerevisiae.FEMS Yeast Res. 2015 Sep;15(6):fov068. doi: 10.1093/femsyr/fov068. Epub 2015 Jul 22. FEMS Yeast Res. 2015. PMID: 26205245 Free PMC article. Review.
Cited by
-
Dynamic Regulation of the 26S Proteasome: From Synthesis to Degradation.Front Mol Biosci. 2019 Jun 7;6:40. doi: 10.3389/fmolb.2019.00040. eCollection 2019. Front Mol Biosci. 2019. PMID: 31231659 Free PMC article. Review.
-
Severe acute respiratory syndrome coronavirus-2 accessory proteins ORF3a and ORF7a modulate autophagic flux and Ca2+ homeostasis in yeast.Front Microbiol. 2023 Apr 3;14:1152249. doi: 10.3389/fmicb.2023.1152249. eCollection 2023. Front Microbiol. 2023. PMID: 37077240 Free PMC article.
-
Mitochondrial volume fraction and translation duration impact mitochondrial mRNA localization and protein synthesis.Elife. 2020 Aug 7;9:e57814. doi: 10.7554/eLife.57814. Elife. 2020. PMID: 32762840 Free PMC article.
-
Coat protein of rice stripe virus enhances autophagy activity through interaction with cytosolic glyceraldehyde-3-phosphate dehydrogenases, a negative regulator of plant autophagy.Stress Biol. 2023 Mar 23;3(1):3. doi: 10.1007/s44154-023-00084-3. Stress Biol. 2023. PMID: 37676568 Free PMC article.
-
Conserved Autophagy Pathway Contributes to Stress Tolerance and Virulence and Differentially Controls Autophagic Flux Upon Nutrient Starvation in Cryptococcus neoformans.Front Microbiol. 2019 Nov 26;10:2690. doi: 10.3389/fmicb.2019.02690. eCollection 2019. Front Microbiol. 2019. PMID: 32038502 Free PMC article.
References
-
- Tsukada M., and Ohsumi Y. (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174 - PubMed
-
- Mizushima N. (2005) The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 12, 1535–1541 - PubMed
-
- Onodera J., and Ohsumi Y. (2004) Ald6p is a preferred target for autophagy in yeast, Saccharomyces cerevisiae. J. Biol. Chem. 279, 16071–16076 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

