Impact of Dietary Resistant Starch on the Human Gut Microbiome, Metaproteome, and Metabolome
- PMID: 29042495
- PMCID: PMC5646248
- DOI: 10.1128/mBio.01343-17
Impact of Dietary Resistant Starch on the Human Gut Microbiome, Metaproteome, and Metabolome
Abstract
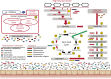
Diet can influence the composition of the human microbiome, and yet relatively few dietary ingredients have been systematically investigated with respect to their impact on the functional potential of the microbiome. Dietary resistant starch (RS) has been shown to have health benefits, but we lack a mechanistic understanding of the metabolic processes that occur in the gut during digestion of RS. Here, we collected samples during a dietary crossover study with diets containing large or small amounts of RS. We determined the impact of RS on the gut microbiome and metabolic pathways in the gut, using a combination of "omics" approaches, including 16S rRNA gene sequencing, metaproteomics, and metabolomics. This multiomics approach captured changes in the abundance of specific bacterial species, proteins, and metabolites after a diet high in resistant starch (HRS), providing key insights into the influence of dietary interventions on the gut microbiome. The combined data showed that a high-RS diet caused an increase in the ratio of Firmicutes to Bacteroidetes, including increases in relative abundances of some specific members of the Firmicutes and concurrent increases in enzymatic pathways and metabolites involved in lipid metabolism in the gut.IMPORTANCE This work was undertaken to obtain a mechanistic understanding of the complex interplay between diet and the microorganisms residing in the intestine. Although it is known that gut microbes play a key role in digestion of the food that we consume, the specific contributions of different microorganisms are not well understood. In addition, the metabolic pathways and resultant products of metabolism during digestion are highly complex. To address these knowledge gaps, we used a combination of molecular approaches to determine the identities of the microorganisms in the gut during digestion of dietary starch as well as the metabolic pathways that they carry out. Together, these data provide a more complete picture of the function of the gut microbiome in digestion, including links between an RS diet and lipid metabolism and novel linkages between specific gut microbes and their metabolites and proteins produced in the gut.
Keywords: gut microbiome; human microbiome; multiomics; resistant starch.
Copyright © 2017 Maier et al.
Figures




References
-
- Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, Louis P, McIntosh F, Johnstone AM, Lobley GE, Parkhill J, Flint HJ. 2011. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5:220–230. doi: 10.1038/ismej.2010.118. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
