Control of growth and gut maturation by HoxD genes and the associated lncRNA Haglr
- PMID: 29042517
- PMCID: PMC5676926
- DOI: 10.1073/pnas.1712511114
Control of growth and gut maturation by HoxD genes and the associated lncRNA Haglr
Abstract
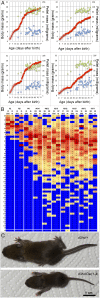
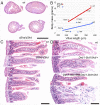
During embryonic development, Hox genes participate in the building of a functional digestive system in metazoans, and genetic conditions involving these genes lead to important, sometimes lethal, growth retardation. Recently, this phenotype was obtained after deletion of Haglr, the Hoxd antisense growth-associated long noncoding RNA (lncRNA) located between Hoxd1 and Hoxd3 In this study, we have analyzed the function of Hoxd genes in delayed growth trajectories by looking at several nested targeted deficiencies of the mouse HoxD cluster. Mutant pups were severely stunted during the suckling period, but many recovered after weaning. After comparing seven distinct HoxD alleles, including CRISPR/Cas9 deletions involving Haglr, we identified Hoxd3 as the critical component for the gut to maintain milk-digestive competence. This essential function could be abrogated by the dominant-negative effect of HOXD10 as shown by a genetic rescue approach, thus further illustrating the importance of posterior prevalence in Hox gene function. A role for the lncRNA Haglr in the control of postnatal growth could not be corroborated.
Keywords: CRISPR-cas9; Hox regulation; digestive system; growth control; lncRNA.
Copyright © 2017 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Rescue of an aggressive female sexual courtship in mice by CRISPR/Cas9 secondary mutation in vivo.BMC Res Notes. 2018 Mar 27;11(1):193. doi: 10.1186/s13104-018-3307-8. BMC Res Notes. 2018. PMID: 29580290 Free PMC article.
-
Long non-coding RNA HOXD-AS1 in cancer.Clin Chim Acta. 2018 Dec;487:197-201. doi: 10.1016/j.cca.2018.10.002. Epub 2018 Oct 2. Clin Chim Acta. 2018. PMID: 30290157 Review.
-
Control of colinearity in AbdB genes of the mouse HoxD complex.Mol Cell. 1998 Jan;1(2):289-300. doi: 10.1016/s1097-2765(00)80029-5. Mol Cell. 1998. PMID: 9659925
-
Multiple enhancers regulate Hoxd genes and the Hotdog LncRNA during cecum budding.Cell Rep. 2013 Oct 17;5(1):137-50. doi: 10.1016/j.celrep.2013.09.002. Epub 2013 Sep 26. Cell Rep. 2013. PMID: 24075990
-
Regulatory landscape of the Hox transcriptome.Int J Dev Biol. 2018;62(11-12):693-704. doi: 10.1387/ijdb.180270em. Int J Dev Biol. 2018. PMID: 30604839 Review.
Cited by
-
Lnc HAGLR Promotes Colon Cancer Progression Through Sponging miR-185-5p and Activating CDK4 and CDK6 in vitro and in vivo.Onco Targets Ther. 2020 Jun 22;13:5913-5925. doi: 10.2147/OTT.S246092. eCollection 2020. Onco Targets Ther. 2020. PMID: 32606801 Free PMC article.
-
SP1-induced HOXD-AS1 promotes malignant progression of cholangiocarcinoma by regulating miR-520c-3p/MYCN.Aging (Albany NY). 2020 Aug 28;12(16):16304-16325. doi: 10.18632/aging.103660. Epub 2020 Aug 28. Aging (Albany NY). 2020. PMID: 32857725 Free PMC article.
-
Hox Proteins in the Regulation of Muscle Development.Front Cell Dev Biol. 2021 Oct 18;9:731996. doi: 10.3389/fcell.2021.731996. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34733846 Free PMC article. Review.
-
FOXA2 controls the cis-regulatory networks of pancreatic cancer cells in a differentiation grade-specific manner.EMBO J. 2019 Oct 15;38(20):e102161. doi: 10.15252/embj.2019102161. Epub 2019 Sep 17. EMBO J. 2019. PMID: 31531882 Free PMC article.
-
Rescue of an aggressive female sexual courtship in mice by CRISPR/Cas9 secondary mutation in vivo.BMC Res Notes. 2018 Mar 27;11(1):193. doi: 10.1186/s13104-018-3307-8. BMC Res Notes. 2018. PMID: 29580290 Free PMC article.
References
-
- Zakany J, Duboule D. A genetic basis for altered sexual behavior in mutant female mice. Curr Biol. 2012;22:1676–1680. - PubMed
-
- Zákány J, Kmita M, Alarcon P, de la Pompa JL, Duboule D. Localized and transient transcription of Hox genes suggests a link between patterning and the segmentation clock. Cell. 2001;106:207–217. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

