Evolutionary genomics of grape (Vitis vinifera ssp. vinifera) domestication
- PMID: 29042518
- PMCID: PMC5676911
- DOI: 10.1073/pnas.1709257114
Evolutionary genomics of grape (Vitis vinifera ssp. vinifera) domestication
Abstract
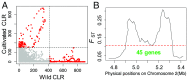
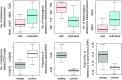
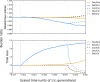
We gathered genomic data from grapes (Vitis vinifera ssp. vinifera), a clonally propagated perennial crop, to address three ongoing mysteries about plant domestication. The first is the duration of domestication; archaeological evidence suggests that domestication occurs over millennia, but genetic evidence indicates that it can occur rapidly. We estimated that our wild and cultivated grape samples diverged ∼22,000 years ago and that the cultivated lineage experienced a steady decline in population size (Ne ) thereafter. The long decline may reflect low-intensity management by humans before domestication. The second mystery is the identification of genes that contribute to domestication phenotypes. In cultivated grapes, we identified candidate-selected genes that function in sugar metabolism, flower development, and stress responses. In contrast, candidate-selected genes in the wild sample were limited to abiotic and biotic stress responses. A genomic region of high divergence corresponded to the sex determination region and included a candidate male sterility factor and additional genes with sex-specific expression. The third mystery concerns the cost of domestication. Annual crops accumulate putatively deleterious variants, in part due to strong domestication bottlenecks. The domestication of perennial crops differs from that of annuals in several ways, including the intensity of bottlenecks, and it is not yet clear if they accumulate deleterious variants. We found that grape accessions contained 5.2% more deleterious variants than wild individuals, and these were more often in a heterozygous state. Using forward simulations, we confirm that clonal propagation leads to the accumulation of recessive deleterious mutations but without decreasing fitness.
Keywords: candidate genes; clonal propagation; deleterious variants; demography; sex determination.
Copyright © 2017 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Tenaillon MI, U’Ren J, Tenaillon O, Gaut BS. Selection versus demography: A multilocus investigation of the domestication process in maize. Mol Biol Evol. 2004;21:1214–1225. - PubMed
-
- Wright SI, et al. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. - PubMed
-
- Meyer RS, Purugganan MD. Evolution of crop species: Genetics of domestication and diversification. Nat Rev Genet. 2013;14:840–852. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

