Structural analysis of PIM1 kinase complexes with ATP-competitive inhibitors
- PMID: 29042609
- PMCID: PMC5645348
- DOI: 10.1038/s41598-017-13557-z
Structural analysis of PIM1 kinase complexes with ATP-competitive inhibitors
Abstract
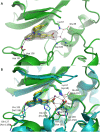
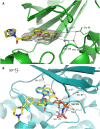
PIM1 is an oncogenic kinase overexpressed in a number of cancers where it correlates with poor prognosis. Several studies demonstrated that inhibition of PIM1 activity is an attractive strategy in fighting overexpressing cancers, while distinct structural features of ATP binding pocket make PIM1 an inviting target for the design of selective inhibitors. To facilitate development of specific PIM1 inhibitors, in this study we report three crystal structures of ATP-competitive inhibitors at the ATP binding pocket of PIM1. Two of the reported structures (CX-4945 and Ro-3306) explain the off-target effect on PIM1 of respectively casein kinase 2 and cyclin-dependent kinase 1 dedicated inhibitors. In turn, the structure with CX-6258 demonstrates a binding mode of a potent, selective inhibitor of PIM1, PIM2, PIM3 and Flt-3 kinases. The consequences of our findings for future inhibitor development are discussed.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
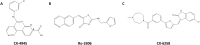

Similar articles
-
Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.Acta Crystallogr D Biol Crystallogr. 2014 Feb;70(Pt 2):392-404. doi: 10.1107/S1399004713028654. Epub 2014 Jan 29. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 24531473
-
Crystal structure of pim1 kinase in complex with a pyrido[4,3-d]pyrimidine derivative suggests a unique binding mode.PLoS One. 2013 Jul 31;8(7):e70358. doi: 10.1371/journal.pone.0070358. Print 2013. PLoS One. 2013. PMID: 23936194 Free PMC article.
-
Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase.Mol Cancer Ther. 2007 Jan;6(1):163-72. doi: 10.1158/1535-7163.MCT-06-0397. Epub 2007 Jan 11. Mol Cancer Ther. 2007. PMID: 17218638
-
Insights from Pim1 structure for anti-cancer drug design.Expert Opin Drug Discov. 2012 Dec;7(12):1177-92. doi: 10.1517/17460441.2012.727394. Epub 2012 Sep 25. Expert Opin Drug Discov. 2012. PMID: 23004574 Review.
-
PIM1 kinase as a target for cancer therapy.Expert Opin Investig Drugs. 2012 Apr;21(4):425-36. doi: 10.1517/13543784.2012.668527. Epub 2012 Mar 4. Expert Opin Investig Drugs. 2012. PMID: 22385334 Review.
Cited by
-
Targeting Echinococcus multilocularis PIM kinase for improving anti-parasitic chemotherapy.PLoS Negl Trop Dis. 2022 Oct 3;16(10):e0010483. doi: 10.1371/journal.pntd.0010483. eCollection 2022 Oct. PLoS Negl Trop Dis. 2022. PMID: 36190997 Free PMC article.
-
Structure-Based Virtual Screening and De Novo Design of PIM1 Inhibitors with Anticancer Activity from Natural Products.Pharmaceuticals (Basel). 2021 Mar 18;14(3):275. doi: 10.3390/ph14030275. Pharmaceuticals (Basel). 2021. PMID: 33803840 Free PMC article.
-
The crystal structure of the protein kinase HIPK2 reveals a unique architecture of its CMGC-insert region.J Biol Chem. 2019 Sep 13;294(37):13545-13559. doi: 10.1074/jbc.RA119.009725. Epub 2019 Jul 24. J Biol Chem. 2019. PMID: 31341017 Free PMC article.
-
Construction of molecular subtype and prognostic model for gastric cancer based on nucleus-encoded mitochondrial genes.Sci Rep. 2024 Nov 18;14(1):28491. doi: 10.1038/s41598-024-78729-0. Sci Rep. 2024. PMID: 39557952 Free PMC article.
-
A systematic review on active sites and functions of PIM-1 protein.Hum Cell. 2022 Mar;35(2):427-440. doi: 10.1007/s13577-021-00656-3. Epub 2022 Jan 9. Hum Cell. 2022. PMID: 35000143
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

