OCT4 impedes cell fate redirection by the melanocyte lineage master regulator MITF in mouse ESCs
- PMID: 29044103
- PMCID: PMC5647326
- DOI: 10.1038/s41467-017-01122-1
OCT4 impedes cell fate redirection by the melanocyte lineage master regulator MITF in mouse ESCs
Abstract
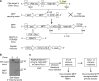
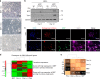
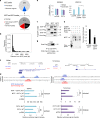
Ectopic expression of lineage master regulators induces transdifferentiation. Whether cell fate transitions can be induced during various developmental stages has not been systemically examined. Here we discover that amongst different developmental stages, mouse embryonic stem cells (mESCs) are resistant to cell fate conversion induced by the melanocyte lineage master regulator MITF. By generating a transgenic system we exhibit that in mESCs, the pluripotency master regulator Oct4, counteracts pro-differentiation induced by Mitf by physical interference with MITF transcriptional activity. We further demonstrate that mESCs must be released from Oct4-maintained pluripotency prior to ectopically induced differentiation. Moreover, Oct4 induction in various differentiated cells represses their lineage identity in vivo. Alongside, chromatin architecture combined with ChIP-seq analysis suggest that Oct4 competes with various lineage master regulators for binding promoters and enhancers. Our analysis reveals pluripotency and transdifferentiation regulatory principles and could open new opportunities in the field of regenerative medicine.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

