Evolution of drift robustness in small populations
- PMID: 29044114
- PMCID: PMC5647343
- DOI: 10.1038/s41467-017-01003-7
Evolution of drift robustness in small populations
Abstract
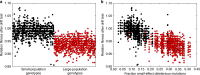
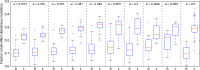
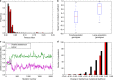
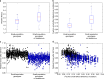
Most mutations are deleterious and cause a reduction in population fitness known as the mutational load. In small populations, weakened selection against slightly-deleterious mutations results in an additional fitness reduction. Many studies have established that populations can evolve a reduced mutational load by evolving mutational robustness, but it is uncertain whether small populations can evolve a reduced susceptibility to drift-related fitness declines. Here, using mathematical modeling and digital experimental evolution, we show that small populations do evolve a reduced vulnerability to drift, or 'drift robustness'. We find that, compared to genotypes from large populations, genotypes from small populations have a decreased likelihood of small-effect deleterious mutations, thus causing small-population genotypes to be drift-robust. We further show that drift robustness is not adaptive, but instead arises because small populations can only maintain fitness on drift-robust fitness peaks. These results have implications for genome evolution in organisms with small effective population sizes.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
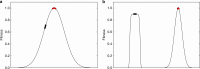
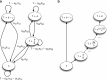
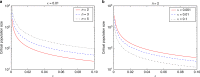
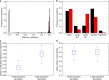
Similar articles
-
Effects of population size and mutation rate on the evolution of mutational robustness.Evolution. 2007 Mar;61(3):666-74. doi: 10.1111/j.1558-5646.2007.00064.x. Evolution. 2007. PMID: 17348929
-
Mapping the Peaks: Fitness Landscapes of the Fittest and the Flattest.Artif Life. 2019 Summer;25(3):250-262. doi: 10.1162/artl_a_00296. Artif Life. 2019. PMID: 31397601
-
Rapid evolutionary escape by large populations from local fitness peaks is likely in nature.Evolution. 2005 Jun;59(6):1175-82. Evolution. 2005. PMID: 16050095
-
Genome size: does bigger mean worse?Curr Biol. 2004 Mar 23;14(6):R233-5. doi: 10.1016/j.cub.2004.02.054. Curr Biol. 2004. PMID: 15043833 Review.
-
Conflicting effects of recombination on the evolvability and robustness in neutrally evolving populations.PLoS Comput Biol. 2022 Nov 21;18(11):e1010710. doi: 10.1371/journal.pcbi.1010710. eCollection 2022 Nov. PLoS Comput Biol. 2022. PMID: 36409763 Free PMC article. Review.
Cited by
-
Cell-line specific RNA editing patterns in Trypanosoma brucei suggest a unique mechanism to generate protein variation in a system intolerant to genetic mutations.Nucleic Acids Res. 2020 Feb 20;48(3):1479-1493. doi: 10.1093/nar/gkz1131. Nucleic Acids Res. 2020. PMID: 31840176 Free PMC article.
-
Admixture on the northern front: population genomics of range expansion in the white-footed mouse (Peromyscus leucopus) and secondary contact with the deer mouse (Peromyscus maniculatus).Heredity (Edinb). 2017 Dec;119(6):447-458. doi: 10.1038/hdy.2017.57. Epub 2017 Sep 13. Heredity (Edinb). 2017. PMID: 28902189 Free PMC article.
-
Macroevolution along developmental lines of least resistance in fly wings.Nat Ecol Evol. 2025 Apr;9(4):639-651. doi: 10.1038/s41559-025-02639-1. Epub 2025 Feb 7. Nat Ecol Evol. 2025. PMID: 39920350 Free PMC article.
-
Low protein expression enhances phenotypic evolvability by intensifying selection on folding stability.Nat Ecol Evol. 2022 Aug;6(8):1155-1164. doi: 10.1038/s41559-022-01797-w. Epub 2022 Jul 7. Nat Ecol Evol. 2022. PMID: 35798838 Free PMC article.
-
Changes in Cell Size and Shape during 50,000 Generations of Experimental Evolution with Escherichia coli.J Bacteriol. 2021 Apr 21;203(10):e00469-20. doi: 10.1128/JB.00469-20. Print 2021 Apr 21. J Bacteriol. 2021. PMID: 33649147 Free PMC article.
References
-
- Crow JF. Some possibilities for measuring selection intensities in man. Human Biology. 1958;30:1–13. - PubMed
-
- Agrawal AF, Whitlock MC. Mutation load: the fitness of individuals in populations where deleterious alleles are abundant. Annual Review of Ecology, Evolution, and Systematics. 2012;43:115–135. doi: 10.1146/annurev-ecolsys-110411-160257. - DOI

