A Machine Learning Assisted, Label-free, Non-invasive Approach for Somatic Reprogramming in Induced Pluripotent Stem Cell Colony Formation Detection and Prediction
- PMID: 29044152
- PMCID: PMC5647349
- DOI: 10.1038/s41598-017-13680-x
A Machine Learning Assisted, Label-free, Non-invasive Approach for Somatic Reprogramming in Induced Pluripotent Stem Cell Colony Formation Detection and Prediction
Abstract
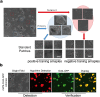
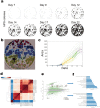
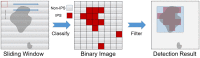
During cellular reprogramming, the mesenchymal-to-epithelial transition is accompanied by changes in morphology, which occur prior to iPSC colony formation. The current approach for detecting morphological changes associated with reprogramming purely relies on human experiences, which involve intensive amounts of upfront training, human error with limited quality control and batch-to-batch variations. Here, we report a time-lapse-based bright-field imaging analysis system that allows us to implement a label-free, non-invasive approach to measure morphological dynamics. To automatically analyse and determine iPSC colony formation, a machine learning-based classification, segmentation, and statistical modelling system was developed to guide colony selection. The system can detect and monitor the earliest cellular texture changes after the induction of reprogramming in human somatic cells on day 7 from the 20-24 day process. Moreover, after determining the reprogramming process and iPSC colony formation quantitatively, a mathematical model was developed to statistically predict the best iPSC selection phase independent of any other resources. All the computational detection and prediction experiments were evaluated using a validation dataset, and biological verification was performed. These algorithm-detected colonies show no significant differences (Pearson Coefficient) in terms of their biological features compared to the manually processed colonies using standard molecular approaches.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

