Matchmaker Exchange
- PMID: 29044468
- PMCID: PMC6016856
- DOI: 10.1002/cphg.50
Matchmaker Exchange
Abstract
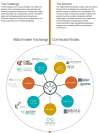
In well over half of the individuals with rare disease who undergo clinical or research next-generation sequencing, the responsible gene cannot be determined. Some reasons for this relatively low yield include unappreciated phenotypic heterogeneity; locus heterogeneity; somatic and germline mosaicism; variants of uncertain functional significance; technically inaccessible areas of the genome; incorrect mode of inheritance investigated; and inadequate communication between clinicians and basic scientists with knowledge of particular genes, proteins, or biological systems. To facilitate such communication and improve the search for patients or model organisms with similar phenotypes and variants in specific candidate genes, we have developed the Matchmaker Exchange (MME). MME was created to establish a federated network connecting databases of genomic and phenotypic data using a common application programming interface (API). To date, seven databases can exchange data using the API (GeneMatcher, PhenomeCentral, DECIPHER, MyGene2, matchbox, Australian Genomics Health Alliance Patient Archive, and Monarch Initiative; the latter included for model organism matching). This article guides usage of the MME for rare disease gene discovery. © 2017 by John Wiley & Sons, Inc.
Keywords: Australian Genomics Health Alliance Patient Archive; DECIPHER; GeneMatcher; MyGene2; PhenomeCentral; candidate genes; matchbox; matchmaker exchange; monarch initiative.
Copyright © 2017 John Wiley and Sons, Inc.
Figures













References
-
- Boycott KM, Rath A, Chong JX, Hartley T, Alkuraya FS, Baynam G, Brookes AJ, Brudno M, Carracedo A, den Dunnen JT, Dyke SOM, Estivill X, Goldblatt J, Gonthier C, Groft SC, Gut I, Hamosh A, Hieter P, Höhn S, Hurles ME, Kaufmann P, Knoppers BM, Krischer JP, Macek M, Jr, Matthijs G, Olry A, Parker S, Paschall J, Philippakis AA, Rehm HL, Robinson PN, Sham PC, Stefanov R, Taruscio D, Unni D, Vanstone MR, Zhang F, Brunner H, Bamshad MJ, Lochmüller H. International Cooperation to Enable the Diagnosis of All Rare Genetic Diseases. Am J Hum Genet. 2017 May 4;100(5):695–705. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

