MicroRNA profiling of human primary macrophages exposed to dengue virus identifies miRNA-3614-5p as antiviral and regulator of ADAR1 expression
- PMID: 29045406
- PMCID: PMC5662241
- DOI: 10.1371/journal.pntd.0005981
MicroRNA profiling of human primary macrophages exposed to dengue virus identifies miRNA-3614-5p as antiviral and regulator of ADAR1 expression
Abstract
Background: Due to the high burden of dengue disease worldwide, a better understanding of the interactions between dengue virus (DENV) and its human host cells is of the utmost importance. Although microRNAs modulate the outcome of several viral infections, their contribution to DENV replication is poorly understood.
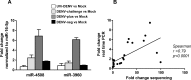
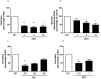
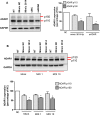
Methods and principal findings: We investigated the microRNA expression profile of primary human macrophages challenged with DENV and deciphered the contribution of microRNAs to infection. To this end, human primary macrophages were challenged with GFP-expressing DENV and sorted to differentiate between truly infected cells (DENV-positive) and DENV-exposed but non-infected cells (DENV-negative cells). The miRNAome was determined by small RNA-Seq analysis and the effect of differentially expressed microRNAs on DENV yield was examined. Five microRNAs were differentially expressed in human macrophages challenged with DENV. Of these, miR-3614-5p was found upregulated in DENV-negative cells and its overexpression reduced DENV infectivity. The cellular targets of miR-3614-5p were identified by liquid chromatography/mass spectrometry and western blot. Adenosine deaminase acting on RNA 1 (ADAR1) was identified as one of the targets of miR-3614-5p and was shown to promote DENV infectivity at early time points post-infection.
Conclusion/significance: Overall, miRNAs appear to play a limited role in DENV replication in primary human macrophages. The miRNAs that were found upregulated in DENV-infected cells did not control the production of infectious virus particles. On the other hand, miR-3614-5p, which was upregulated in DENV-negative macrophages, reduced DENV infectivity and regulated ADAR1 expression, a protein that facilitates viral replication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013. April 25;496(7446):504–7. doi: 10.1038/nature12060 - DOI - PMC - PubMed
-
- World Health Organization. Dengue: guidelines for diagnosis, treatment, prevention, and control. Spec Program Res Train Trop Dis. 2009;147. - PubMed
-
- Guzman MG, Alvarez M, Halstead SB. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: An historical perspective and role of antibody-dependent enhancement of infection. Arch Virol. 2013;158(7):1445–59. doi: 10.1007/s00705-013-1645-3 - DOI - PubMed
-
- Halstead SB. Licensed dengue vaccine: Public health conundrum and scientific challenge. Am J Trop Med Hyg. 2016;95(4):741–5. doi: 10.4269/ajtmh.16-0222 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

