Novel drug targets in cell wall biosynthesis exploited by gene disruption in Pseudomonas aeruginosa
- PMID: 29045498
- PMCID: PMC5646862
- DOI: 10.1371/journal.pone.0186801
Novel drug targets in cell wall biosynthesis exploited by gene disruption in Pseudomonas aeruginosa
Abstract
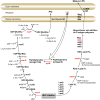
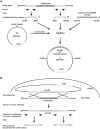
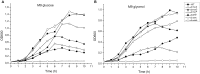
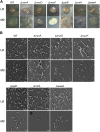
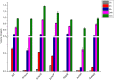
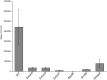
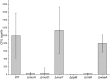
For clinicians, Pseudomonas aeruginosa is a nightmare pathogen that is one of the top three causes of opportunistic human infections. Therapy of P. aeruginosa infections is complicated due to its natural high intrinsic resistance to antibiotics. Active efflux and decreased uptake of drugs due to cell wall/membrane permeability appear to be important issues in the acquired antibiotic tolerance mechanisms. Bacterial cell wall biosynthesis enzymes have been shown to be essential for pathogenicity of Gram-negative bacteria. However, the role of these targets in virulence has not been identified in P. aeruginosa. Here, we report knockout (k.o) mutants of six cell wall biosynthesis targets (murA, PA4450; murD, PA4414; murF, PA4416; ppiB, PA1793; rmlA, PA5163; waaA, PA4988) in P. aeruginosa PAO1, and characterized these in order to find out whether these genes and their products contribute to pathogenicity and virulence of P. aeruginosa. Except waaA k.o, deletion of cell wall biosynthesis targets significantly reduced growth rate in minimal medium compared to the parent strain. The k.o mutants showed exciting changes in cell morphology and colonial architectures. Remarkably, ΔmurF cells became grossly enlarged. Moreover, the mutants were also attenuated in vivo in a mouse infection model except ΔmurF and ΔwaaA and proved to be more sensitive to macrophage-mediated killing than the wild-type strain. Interestingly, the deletion of the murA gene resulted in loss of virulence activity in mice, and the virulence was restored in a plant model by unknown mechanism. This study demonstrates that cell wall targets contribute significantly to intracellular survival, in vivo growth, and pathogenesis of P. aeruginosa. In conclusion, these findings establish a link between cell wall targets and virulence of P. aeruginosa and thus may lead to development of novel drugs for the treatment of P. aeruginosa infection.
Conflict of interest statement
Figures

Similar articles
-
A Genetic Screen Reveals Novel Targets to Render Pseudomonas aeruginosa Sensitive to Lysozyme and Cell Wall-Targeting Antibiotics.Front Cell Infect Microbiol. 2017 Mar 1;7:59. doi: 10.3389/fcimb.2017.00059. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28299285 Free PMC article.
-
The Small RNA ErsA Plays a Role in the Regulatory Network of Pseudomonas aeruginosa Pathogenicity in Airway Infections.mSphere. 2020 Oct 14;5(5):e00909-20. doi: 10.1128/mSphere.00909-20. mSphere. 2020. PMID: 33055260 Free PMC article.
-
Impact of Peptidoglycan Recycling Blockade and Expression of Horizontally Acquired β-Lactamases on Pseudomonas aeruginosa Virulence.Microbiol Spectr. 2022 Feb 23;10(1):e0201921. doi: 10.1128/spectrum.02019-21. Epub 2022 Feb 16. Microbiol Spectr. 2022. PMID: 35171032 Free PMC article.
-
Pseudomonas aeruginosa: targeting cell-wall metabolism for new antibacterial discovery and development.Future Med Chem. 2016 Jun;8(9):975-92. doi: 10.4155/fmc-2016-0017. Epub 2016 May 26. Future Med Chem. 2016. PMID: 27228070 Review.
-
The intrinsic resistome of Pseudomonas aeruginosa to β-lactams.Virulence. 2011 Mar-Apr;2(2):144-6. doi: 10.4161/viru.2.2.15014. Epub 2011 Mar 1. Virulence. 2011. PMID: 21304266 Review.
Cited by
-
Toward a structome of Acinetobacter baumannii drug targets.Protein Sci. 2020 Mar;29(3):789-802. doi: 10.1002/pro.3826. Epub 2020 Jan 20. Protein Sci. 2020. PMID: 31930600 Free PMC article.
-
Profiling the susceptibility of Pseudomonas aeruginosa strains from acute and chronic infections to cell-wall-targeting immune proteins.Sci Rep. 2019 Mar 5;9(1):3575. doi: 10.1038/s41598-019-40440-w. Sci Rep. 2019. PMID: 30837659 Free PMC article.
-
A Systematic Strategy to Find Potential Therapeutic Targets for Pseudomonas aeruginosa Using Integrated Computational Models.Front Mol Biosci. 2021 Sep 20;8:728129. doi: 10.3389/fmolb.2021.728129. eCollection 2021. Front Mol Biosci. 2021. PMID: 34616771 Free PMC article.
-
Exploring nagZ as a virulence biomarker and treatment target in Enterobacter cloacae.BMC Microbiol. 2025 Jan 9;25(1):10. doi: 10.1186/s12866-024-03718-2. BMC Microbiol. 2025. PMID: 39789468 Free PMC article.
-
Toad's survivability and soil microbiome alterations impacted via individual abundance.Biol Futur. 2025 Sep;76(3):399-411. doi: 10.1007/s42977-025-00261-7. Epub 2025 Jun 6. Biol Futur. 2025. PMID: 40478395
References
-
- Bodey GP, Bolivar R, Fainstein V, Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983;5(2):279–313. Epub 1983/03/01. . - PubMed
-
- Amini S, Hottes AK, Smith LE, Tavazoie S. Fitness landscape of antibiotic tolerance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2011;7(10):e1002298 Epub 2011/10/27. doi: 10.1371/journal.ppat.1002298 - DOI - PMC - PubMed
-
- Mulcahy H, Charron-Mazenod L, Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008;4(11):e1000213 Epub 2008/11/22. doi: 10.1371/journal.ppat.1000213 - DOI - PMC - PubMed
-
- Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis. 2002;34(5):634–40. Epub 2002/02/02. doi: 10.1086/338782 . - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

