Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients
- PMID: 29045505
- PMCID: PMC6248757
- DOI: 10.1093/annonc/mdx542
Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients
Abstract
Background: Detection of circulating tumor DNA can be limited due to their relative scarcity in circulation, particularly while patients are actively undergoing therapy. Exosomes provide a vehicle through which cancer-specific material can be enriched from the compendium of circulating non-neoplastic tissue-derived nucleic acids. We carried out a comprehensive profiling of the pancreatic ductal adenocarcinoma (PDAC) exosomal 'surfaceome' in order to identify surface proteins that will render liquid biopsies amenable to cancer-derived exosome enrichment for downstream molecular profiling.
Patients and methods: Surface exosomal proteins were profiled in 13 human PDAC and 2 non-neoplastic cell lines by liquid chromatography-mass spectrometry. A total of 173 prospectively collected blood samples from 103 PDAC patients underwent exosome isolation. Droplet digital PCR was used on 74 patients (136 total exosome samples) to determine baseline KRAS mutation call rates while patients were on therapy. PDAC-specific exosome capture was then carried out on additional 29 patients (37 samples) using an antibody cocktail directed against selected proteins, followed by droplet digital PCR analysis. Exosomal DNA in a PDAC patient resistant to therapy were profiled using a molecular barcoded, targeted sequencing panel to determine the utility of enriched nucleic acid material for comprehensive molecular analysis.
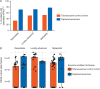
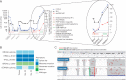
Results: Proteomic analysis of the exosome 'surfaceome' revealed multiple PDAC-specific biomarker candidates: CLDN4, EPCAM, CD151, LGALS3BP, HIST2H2BE, and HIST2H2BF. KRAS mutations in total exosomes were detected in 44.1% of patients undergoing active therapy compared with 73.0% following exosome capture using the selected biomarkers. Enrichment of exosomal cargo was amenable to molecular profiling, elucidating a putative mechanism of resistance to PARP inhibitor therapy in a patient harboring a BRCA2 mutation.
Conclusion: Exosomes provide unique opportunities in the context of liquid biopsies for enrichment of tumor-specific material in circulation. We present a comprehensive surfaceome characterization of PDAC exosomes which allows for capture and molecular profiling of tumor-derived DNA.
Keywords: exosomes; liquid biopsies; next-generation sequencing; pancreatic cancer; proteomics.
© The Author 2017. Published by Oxford University Press on behalf of the European Society for Medical Oncology. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures

References
-
- Bailey P, Chang DK, Nones K. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016; 531: 47–52. http://dx.doi.org/10.1038/nature16965 - DOI - PubMed
-
- Waddell N, Pajic M, Patch AM. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015; 518: 495–501. http://dx.doi.org/10.1038/nature14169 - DOI - PMC - PubMed
-
- San Lucas FA, Allenson K, Bernard V. et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann Oncol 2016; 27: 635–664. http://dx.doi.org/10.1093/annonc/mdv604 - DOI - PMC - PubMed
-
- Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer 2016; 16: 582–598. http://dx.doi.org/10.1038/nrc.2016.73 - DOI - PubMed
-
- Costa-Silva B, Aiello NM, Ocean AJ. et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol 2015; 17: 816–826. http://dx.doi.org/10.1038/ncb3169 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

