Evaluation of variable selection methods for random forests and omics data sets
- PMID: 29045534
- PMCID: PMC6433899
- DOI: 10.1093/bib/bbx124
Evaluation of variable selection methods for random forests and omics data sets
Abstract
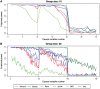
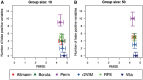
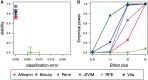
Machine learning methods and in particular random forests are promising approaches for prediction based on high dimensional omics data sets. They provide variable importance measures to rank predictors according to their predictive power. If building a prediction model is the main goal of a study, often a minimal set of variables with good prediction performance is selected. However, if the objective is the identification of involved variables to find active networks and pathways, approaches that aim to select all relevant variables should be preferred. We evaluated several variable selection procedures based on simulated data as well as publicly available experimental methylation and gene expression data. Our comparison included the Boruta algorithm, the Vita method, recurrent relative variable importance, a permutation approach and its parametric variant (Altmann) as well as recursive feature elimination (RFE). In our simulation studies, Boruta was the most powerful approach, followed closely by the Vita method. Both approaches demonstrated similar stability in variable selection, while Vita was the most robust approach under a pure null model without any predictor variables related to the outcome. In the analysis of the different experimental data sets, Vita demonstrated slightly better stability in variable selection and was less computationally intensive than Boruta. In conclusion, we recommend the Boruta and Vita approaches for the analysis of high-dimensional data sets. Vita is considerably faster than Boruta and thus more suitable for large data sets, but only Boruta can also be applied in low-dimensional settings.
Keywords: feature selection; high dimensional data; machine learning; random forest; relevant variables.
© The Author 2017. Published by Oxford University Press.
Figures



References
-
- Breiman L. Random forests. Mach Learn 2001;45:5–32.
-
- Szymczak S, Biernacka JM, Cordell HJ, et al. Machine learning in genome-wide association studies. Genet Epidemiol 2009;33:S51–7. - PubMed
-
- Alexe G, Monaco J, Doyle S, et al. Towards improved cancer diagnosis and prognosis using analysis of gene expression data and computer aided imaging. Exp Biol Med 2009;234:860–79. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

