CTLA-4+PD-1- Memory CD4+ T Cells Critically Contribute to Viral Persistence in Antiretroviral Therapy-Suppressed, SIV-Infected Rhesus Macaques
- PMID: 29045906
- PMCID: PMC5679306
- DOI: 10.1016/j.immuni.2017.09.018
CTLA-4+PD-1- Memory CD4+ T Cells Critically Contribute to Viral Persistence in Antiretroviral Therapy-Suppressed, SIV-Infected Rhesus Macaques
Abstract
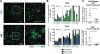
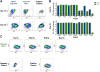
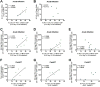
Antiretroviral therapy (ART) suppresses viral replication in HIV-infected individuals but does not eliminate the reservoir of latently infected cells. Recent work identified PD-1+ follicular helper T (Tfh) cells as an important cellular compartment for viral persistence. Here, using ART-treated, SIV-infected rhesus macaques, we show that CTLA-4+PD-1- memory CD4+ T cells, which share phenotypic markers with regulatory T cells, were enriched in SIV DNA in blood, lymph nodes (LN), spleen, and gut, and contained replication-competent and infectious virus. In contrast to PD-1+ Tfh cells, SIV-enriched CTLA-4+PD-1- CD4+ T cells were found outside the B cell follicle of the LN, predicted the size of the persistent viral reservoir during ART, and significantly increased their contribution to the SIV reservoir with prolonged ART-mediated viral suppression. We have shown that CTLA-4+PD-1- memory CD4+ T cells are a previously unrecognized component of the SIV and HIV reservoir that should be therapeutically targeted for a functional HIV-1 cure.
Keywords: CTLA-4; HIV; PD-1; SIV; T follicular helper cells; animal models; co-inhibitory receptors; latent viral reservoir; regulatory T cells.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors have declared that no conflict of interest exists.
Figures




Similar articles
-
Bone Marrow-Derived CD4+ T Cells Are Depleted in Simian Immunodeficiency Virus-Infected Macaques and Contribute to the Size of the Replication-Competent Reservoir.J Virol. 2018 Dec 10;93(1):e01344-18. doi: 10.1128/JVI.01344-18. Print 2019 Jan 1. J Virol. 2018. PMID: 30305357 Free PMC article.
-
CTLA-4 and PD-1 dual blockade induces SIV reactivation without control of rebound after antiretroviral therapy interruption.Nat Med. 2020 Apr;26(4):519-528. doi: 10.1038/s41591-020-0782-y. Epub 2020 Mar 16. Nat Med. 2020. PMID: 32284611 Free PMC article.
-
Initiation of Antiretroviral Therapy Restores CD4+ T Memory Stem Cell Homeostasis in Simian Immunodeficiency Virus-Infected Macaques.J Virol. 2016 Jul 11;90(15):6699-6708. doi: 10.1128/JVI.00492-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27170752 Free PMC article.
-
Brain macrophages harbor latent, infectious simian immunodeficiency virus.AIDS. 2019 Dec 1;33 Suppl 2(Suppl 2):S181-S188. doi: 10.1097/QAD.0000000000002269. AIDS. 2019. PMID: 31789817 Free PMC article. Review.
-
HIV Persistence in Adipose Tissue Reservoirs.Curr HIV/AIDS Rep. 2018 Feb;15(1):60-71. doi: 10.1007/s11904-018-0378-z. Curr HIV/AIDS Rep. 2018. PMID: 29423731 Free PMC article. Review.
Cited by
-
The Role of Tissue Resident Memory CD4 T Cells in Herpes Simplex Viral and HIV Infection.Viruses. 2021 Feb 25;13(3):359. doi: 10.3390/v13030359. Viruses. 2021. PMID: 33668777 Free PMC article. Review.
-
The Complex Dysregulations of CD4 T Cell Subtypes in HIV Infection.Int J Mol Sci. 2024 Jul 9;25(14):7512. doi: 10.3390/ijms25147512. Int J Mol Sci. 2024. PMID: 39062756 Free PMC article. Review.
-
HIV Rebound Is Predominantly Fueled by Genetically Identical Viral Expansions from Diverse Reservoirs.Cell Host Microbe. 2019 Sep 11;26(3):347-358.e7. doi: 10.1016/j.chom.2019.08.003. Epub 2019 Aug 27. Cell Host Microbe. 2019. PMID: 31471273 Free PMC article.
-
Silence, escape and survival drive the persistence of HIV.Nature. 2023 Feb;614(7947):236-237. doi: 10.1038/d41586-022-04492-9. Nature. 2023. PMID: 36599993 No abstract available.
-
Tissue memory CD4+ T cells expressing IL-7 receptor-alpha (CD127) preferentially support latent HIV-1 infection.PLoS Pathog. 2020 Apr 30;16(4):e1008450. doi: 10.1371/journal.ppat.1008450. eCollection 2020 Apr. PLoS Pathog. 2020. PMID: 32353080 Free PMC article.
References
-
- Amara RR, Villinger F, Altman JD, Lydy SL, O’Neil SP, Staprans SI, Montefiori DC, Xu Y, Herndon JG, Wyatt LS, et al. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science. 2001;292:69–74. - PubMed
-
- Anderson A, Martens CL, Hendrix R, Stempora LL, Miller WP, Hamby K, Russell M, Strobert E, Blazar BR, Pearson TC, et al. Expanded nonhuman primate tregs exhibit a unique gene expression signature and potently downregulate alloimmune responses. American Journal of Transplantation. 2008;8:2252–2264. - PMC - PubMed
-
- Autran B, Carcelain G, Li TS, Blanc C, Mathez D, Tubiana R, Katlama C, Debre P, Leibowitch J. Positive effects of combined antiretroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science. 1997;277:112–116. - PubMed
-
- Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, Corpataux JM, de Leval L, Pantaleo G, Perreau M. PD-1 and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med 2016 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

