Detection of Plasmodium Species by High-Resolution Melt Analysis of DNA from Blood Smears Acquired in Southwestern Uganda
- PMID: 29046412
- PMCID: PMC5744206
- DOI: 10.1128/JCM.01060-17
Detection of Plasmodium Species by High-Resolution Melt Analysis of DNA from Blood Smears Acquired in Southwestern Uganda
Abstract
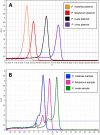
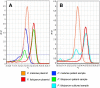
Microscopic diagnosis of malaria using Giemsa-stained blood smears is the standard of care in resource-limited settings. These smears represent a potential source of DNA for PCR testing to confirm Plasmodium infections or for epidemiological studies of archived samples. Therefore, we assessed the use of DNA extracts from stained blood smears for the detection of Plasmodium species using real-time PCR. We extracted DNA from archived blood smears and corresponding red blood cell pellets collected from asymptomatic children in southwestern Uganda in 2010. We then performed real-time PCR followed by high-resolution melting (HRM) to identify Plasmodium species, and we compared our results to those of microscopy. We analyzed a total of 367 blood smears and corresponding red blood cell pellets, including 185 smears (50.4%) that were positive by microscopy. Compared to microscopy, PCR-HRM analysis of smear DNA had a sensitivity of 93.0% (95% confidence interval [CI], 88.2 to 96.2%) and a specificity of 96.7% (95% CI, 93.0 to 98.8%), and PCR-HRM analysis of pellet DNA had a sensitivity of 100.0% (95% CI, 98.0 to 100.0%) and a specificity of 94.0% (95% CI, 89.4 to 96.9%). Identification of positive PCR-HRM results to the species level revealed Plasmodium falciparum (92.0%), Plasmodium ovale (5.6%), and Plasmodium malariae (2.4%). PCR-HRM analysis of DNA extracts from Giemsa-stained thick blood smears or corresponding blood pellets had high sensitivity and specificity for malaria diagnosis, compared to microscopy. Therefore, blood smears can provide an adequate source of DNA for confirmation of Plasmodium species infections and can be used for retrospective genetic studies.
Keywords: PCR; diagnostics; high-resolution melting; malaria.
Copyright © 2017 American Society for Microbiology.
Figures

Similar articles
-
Low sensitivity of nested PCR using Plasmodium DNA extracted from stained thick blood smears: an epidemiological retrospective study among subjects with low parasitaemia in an endemic area of the Brazilian Amazon region.Malar J. 2004 Mar 31;3:8. doi: 10.1186/1475-2875-3-8. Malar J. 2004. PMID: 15056392 Free PMC article.
-
Simple Real-Time PCR and Amplicon Sequencing Method for Identification of Plasmodium Species in Human Whole Blood.J Clin Microbiol. 2015 Jul;53(7):2251-7. doi: 10.1128/JCM.00542-15. Epub 2015 May 13. J Clin Microbiol. 2015. PMID: 25972416 Free PMC article.
-
Detection of malaria with light microscopy and Nested polymerase chain reaction (Nested PCR) methods in peripheral blood expansions and investigation of the genetic diversity of Plasmodium species by 18S rRNA gene in Southeast of Iran.Microb Pathog. 2019 Dec;137:103782. doi: 10.1016/j.micpath.2019.103782. Epub 2019 Oct 7. Microb Pathog. 2019. PMID: 31600540
-
High-resolution melt curve analysis: A real-time based multipurpose approach for diagnosis and epidemiological investigations of parasitic infections.Comp Immunol Microbiol Infect Dis. 2019 Dec;67:101364. doi: 10.1016/j.cimid.2019.101364. Epub 2019 Sep 28. Comp Immunol Microbiol Infect Dis. 2019. PMID: 31590033 Review.
-
Emerging nucleic acid-based tests for point-of-care detection of malaria.Am J Trop Med Hyg. 2012 Aug;87(2):223-230. doi: 10.4269/ajtmh.2012.11-0685. Am J Trop Med Hyg. 2012. PMID: 22855751 Free PMC article. Review.
Cited by
-
Molecular identification and anti-malarial drug resistance profile of Plasmodium falciparum from patients attending Kisoro Hospital, southwestern Uganda.Malar J. 2022 Jan 15;21(1):21. doi: 10.1186/s12936-021-04023-3. Malar J. 2022. PMID: 35033082 Free PMC article.
-
Antimalarial resistance risk in Mozambique detected by a novel quadruplex droplet digital PCR assay.Antimicrob Agents Chemother. 2024 Jul 9;68(7):e0034624. doi: 10.1128/aac.00346-24. Epub 2024 May 21. Antimicrob Agents Chemother. 2024. PMID: 38771031 Free PMC article.
-
A new high-resolution melting analysis for the detection and identification of Plasmodium in human and Anopheles vectors of malaria.Sci Rep. 2019 Feb 8;9(1):1674. doi: 10.1038/s41598-018-36515-9. Sci Rep. 2019. PMID: 30737420 Free PMC article.
-
Distinction of Plasmodium ovale wallikeri and Plasmodium ovale curtisi using quantitative Polymerase Chain Reaction with High Resolution Melting revelation.Sci Rep. 2018 Jan 10;8(1):300. doi: 10.1038/s41598-017-18026-1. Sci Rep. 2018. PMID: 29321578 Free PMC article.
-
Permethrin-treated baby wraps for the prevention of malaria: results of a randomized controlled pilot study in rural Uganda.Malar J. 2022 Feb 23;21(1):63. doi: 10.1186/s12936-022-04086-w. Malar J. 2022. PMID: 35197060 Free PMC article. Clinical Trial.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

