Resistance of Major Histocompatibility Complex Class B (MHC-B) to Nef-Mediated Downregulation Relative to that of MHC-A Is Conserved among Primate Lentiviruses and Influences Antiviral T Cell Responses in HIV-1-Infected Individuals
- PMID: 29046444
- PMCID: PMC5730772
- DOI: 10.1128/JVI.01409-17
Resistance of Major Histocompatibility Complex Class B (MHC-B) to Nef-Mediated Downregulation Relative to that of MHC-A Is Conserved among Primate Lentiviruses and Influences Antiviral T Cell Responses in HIV-1-Infected Individuals
Abstract
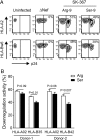
Patient-derived HIV-1 subtype B Nef clones downregulate HLA-A more efficiently than HLA-B. However, it remains unknown whether this property is common to Nef proteins across primate lentiviruses and how antiviral immune responses may be affected. We examined 263 Nef clones from diverse primate lentiviruses including different pandemic HIV-1 group M subtypes for their ability to downregulate major histocompatibility complex class A (MHC-A) and MHC-B from the cell surface. Though lentiviral Nef proteins differed markedly in their absolute MHC-A and MHC-B downregulation abilities, all lentiviral Nef lineages downregulated MHC-A, on average, 11 to 32% more efficiently than MHC-B. Nef genotype/phenotype analyses in a cohort of HIV-1 subtype C-infected patients (n = 168), together with site-directed mutagenesis, revealed Nef position 9 as a subtype-specific determinant of differential HLA-A versus HLA-B downregulation activity. Nef clones harboring nonconsensus variants at codon 9 downregulated HLA-B (though not HLA-A) significantly better than those harboring the consensus sequence at this site, resulting in reduced recognition of infected target cells by HIV-1-specific CD8+ effector cells in vitro Among persons expressing protective HLA class I alleles, carriage of Nef codon 9 variants was also associated with reduced ex vivo HIV-specific T cell responses. Our results demonstrate that Nef's inferior ability to downregulate MHC-B compared to that of MHC-A is conserved across primate lentiviruses and suggest that this property influences antiviral cellular immune responses.IMPORTANCE Primate lentiviruses encode the Nef protein that plays an essential role in establishing persistent infection in their respective host species. Nef interacts with the cytoplasmic region of MHC-A and MHC-B molecules and downregulates them from the infected cell surface to escape recognition by host cellular immunity. Using a panel of Nef alleles isolated from diverse primate lentiviruses including pandemic HIV-1 group M subtypes, we demonstrate that Nef proteins across all lentiviral lineages downregulate MHC-A approximately 20% more effectively than MHC-B. We further identify a naturally polymorphic site at Nef position 9 that contributes to the MHC-B downregulation function in HIV-1 subtype C and show that carriage of Nef variants with enhanced MHC-B downregulation ability is associated with reduced breadth and magnitude of MHC-B-restricted cellular immune responses in HIV-infected individuals. Our study underscores an evolutionarily conserved interaction between lentiviruses and primate immune systems that may contribute to pathogenesis.
Keywords: HLA; Nef; human immunodeficiency virus; immune evasion; lentiviruses.
Copyright © 2017 American Society for Microbiology.
Figures


Similar articles
-
Relative Resistance of HLA-B to Downregulation by Naturally Occurring HIV-1 Nef Sequences.mBio. 2016 Jan 19;7(1):e01516-15. doi: 10.1128/mBio.01516-15. mBio. 2016. PMID: 26787826 Free PMC article.
-
HLA Class I Downregulation by HIV-1 Variants from Subtype C Transmission Pairs.J Virol. 2018 Mar 14;92(7):e01633-17. doi: 10.1128/JVI.01633-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29321314 Free PMC article.
-
Antibodies and lentiviruses that specifically recognize a T cell epitope derived from HIV-1 Nef protein and presented by HLA-C.J Immunol. 2010 Dec 15;185(12):7623-32. doi: 10.4049/jimmunol.1001561. Epub 2010 Nov 12. J Immunol. 2010. PMID: 21076072
-
HIV's evasion of the cellular immune response.Immunol Rev. 1999 Apr;168:65-74. doi: 10.1111/j.1600-065x.1999.tb01283.x. Immunol Rev. 1999. PMID: 10399065 Review.
-
HIV-1 Nef: a master manipulator of the membrane trafficking machinery mediating immune evasion.Biochim Biophys Acta. 2015 Apr;1850(4):733-41. doi: 10.1016/j.bbagen.2015.01.003. Epub 2015 Jan 10. Biochim Biophys Acta. 2015. PMID: 25585010 Review.
Cited by
-
SARS-CoV-2 accessory protein ORF8 is secreted extracellularly as a glycoprotein homodimer.J Biol Chem. 2022 Mar;298(3):101724. doi: 10.1016/j.jbc.2022.101724. Epub 2022 Feb 11. J Biol Chem. 2022. PMID: 35157849 Free PMC article.
-
The Persistence of HIV Diversity, Transcription, and Nef Protein in Kaposi's Sarcoma Tumors during Antiretroviral Therapy.Viruses. 2022 Dec 13;14(12):2774. doi: 10.3390/v14122774. Viruses. 2022. PMID: 36560778 Free PMC article.
-
Host-virus interaction and viral evasion.Cell Biol Int. 2021 Jun;45(6):1124-1147. doi: 10.1002/cbin.11565. Epub 2021 Feb 19. Cell Biol Int. 2021. PMID: 33533523 Free PMC article. Review.
-
The Evolutionary Arms Race between Virus and NK Cells: Diversity Enables Population-Level Virus Control.Viruses. 2019 Oct 17;11(10):959. doi: 10.3390/v11100959. Viruses. 2019. PMID: 31627371 Free PMC article. Review.
-
HLA-B*57:01 Complexed to a CD8 T-Cell Epitope from the HSV-2 ICP22 Protein Binds NK and T Cells through KIR3DL1.Viruses. 2022 May 11;14(5):1019. doi: 10.3390/v14051019. Viruses. 2022. PMID: 35632760 Free PMC article.
References
-
- Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O'Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DM, Vine S, Addo MM, Allen TM, et al. . 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557. doi:10.1126/science.1195271. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

