Human alternative Klotho mRNA is a nonsense-mediated mRNA decay target inefficiently spliced in renal disease
- PMID: 29046474
- PMCID: PMC5846909
- DOI: 10.1172/jci.insight.94375
Human alternative Klotho mRNA is a nonsense-mediated mRNA decay target inefficiently spliced in renal disease
Abstract
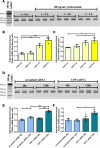
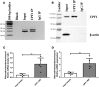
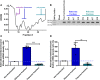
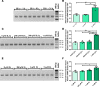
Klotho is a renal protein involved in phosphate homeostasis, which is downregulated in renal disease. It has long been considered an antiaging factor. Two Klotho gene transcripts are thought to encode membrane-bound and secreted Klotho. Indeed, soluble Klotho is detectable in bodily fluids, but the relative contributions of Klotho secretion and of membrane-bound Klotho shedding are unknown. Recent advances in RNA surveillance reveal that premature termination codons, as present in alternative Klotho mRNA (for secreted Klotho), prime mRNAs for degradation by nonsense-mediated mRNA decay (NMD). Disruption of NMD led to accumulation of alternative Klotho mRNA, indicative of normally continuous degradation. RNA IP for NMD core factor UPF1 resulted in enrichment for alternative Klotho mRNA, which was also not associated with polysomes, indicating no active protein translation. Alternative Klotho mRNA transcripts colocalized with some P bodies, where NMD transcripts are degraded. Moreover, we could not detect secreted Klotho in vitro. These results suggest that soluble Klotho is likely cleaved membrane-bound Klotho only. Furthermore, we found that, especially in acute kidney injury, splicing of the 2 mRNA transcripts is dysregulated, which was recapitulated by various noxious stimuli in vitro. This likely constitutes a novel mechanism resulting in the downregulation of membrane-bound Klotho.
Conflict of interest statement
Figures




Similar articles
-
p.Val19Glyfs*21 and p.Leu228* variants in the survival of motor neuron 1 trigger nonsense-mediated mRNA decay causing the SMN1 PTC+ transcripts degradation.Mutat Res. 2017 Dec;806:31-38. doi: 10.1016/j.mrfmmm.2017.09.005. Epub 2017 Sep 15. Mutat Res. 2017. PMID: 28950212
-
Biochemical characterization of the RNA helicase UPF1 involved in nonsense-mediated mRNA decay.Methods Enzymol. 2012;511:255-74. doi: 10.1016/B978-0-12-396546-2.00012-7. Methods Enzymol. 2012. PMID: 22713324
-
Upf proteins: highly conserved factors involved in nonsense mRNA mediated decay.Mol Biol Rep. 2018 Feb;45(1):39-55. doi: 10.1007/s11033-017-4139-7. Epub 2017 Dec 27. Mol Biol Rep. 2018. PMID: 29282598 Review.
-
Defining nonsense-mediated mRNA decay intermediates in human cells.Methods. 2019 Feb 15;155:68-76. doi: 10.1016/j.ymeth.2018.12.005. Epub 2018 Dec 19. Methods. 2019. PMID: 30576707 Free PMC article. Review.
-
An alternative UPF1 isoform drives conditional remodeling of nonsense-mediated mRNA decay.EMBO J. 2022 May 16;41(10):e108898. doi: 10.15252/embj.2021108898. Epub 2022 Apr 11. EMBO J. 2022. PMID: 35403729 Free PMC article.
Cited by
-
Klotho: A Major Shareholder in Vascular Aging Enterprises.Int J Mol Sci. 2019 Sep 19;20(18):4637. doi: 10.3390/ijms20184637. Int J Mol Sci. 2019. PMID: 31546756 Free PMC article. Review.
-
Is there a role in acute kidney injury for FGF23 and Klotho?Clin Kidney J. 2023 Apr 25;16(10):1555-1562. doi: 10.1093/ckj/sfad093. eCollection 2023 Oct. Clin Kidney J. 2023. PMID: 37779849 Free PMC article. Review.
-
α-Klotho's effects on mineral homeostasis are fibroblast growth factor-23 dependent.Curr Opin Nephrol Hypertens. 2018 Jul;27(4):229-235. doi: 10.1097/MNH.0000000000000415. Curr Opin Nephrol Hypertens. 2018. PMID: 29851418 Free PMC article. Review.
-
FGF23 expression is stimulated in transgenic α-Klotho longevity mouse model.JCI Insight. 2019 Dec 5;4(23):e132820. doi: 10.1172/jci.insight.132820. JCI Insight. 2019. PMID: 31801907 Free PMC article.
-
Pathobiology of the Klotho Antiaging Protein and Therapeutic Considerations.Front Aging. 2022 Jul 12;3:931331. doi: 10.3389/fragi.2022.931331. eCollection 2022. Front Aging. 2022. PMID: 35903083 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

