Synaptopodin is upregulated by IL-13 in eosinophilic esophagitis and regulates esophageal epithelial cell motility and barrier integrity
- PMID: 29046486
- PMCID: PMC5846900
- DOI: 10.1172/jci.insight.96789
Synaptopodin is upregulated by IL-13 in eosinophilic esophagitis and regulates esophageal epithelial cell motility and barrier integrity
Abstract
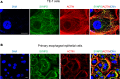
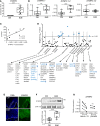
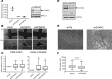
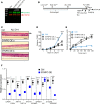
Eosinophilic esophagitis (EoE) is an allergic inflammatory disease of the esophagus mediated by an IL-13-driven epithelial cell transcriptional program. Herein, we show that the cytoskeletal protein synaptopodin (SYNPO), previously associated with podocytes, is constitutively expressed in esophageal epithelium and induced during allergic inflammation. In addition, we show that the SYNPO gene is transcriptionally and epigenetically regulated by IL-13 in esophageal epithelial cells. SYNPO was expressed in the basal layer of homeostatic esophageal epithelium, colocalized with actin filaments, and expanded into the suprabasal epithelium in EoE patients, where expression was elevated 25-fold compared with control individuals. The expression level of SYNPO in esophageal biopsies correlated with esophageal eosinophil density and was improved following anti-IL-13 treatment in EoE patients. In esophageal epithelial cells, SYNPO gene silencing reduced epithelial motility in a wound healing model, whereas SYNPO overexpression impaired epithelial barrier integrity and reduced esophageal differentiation. Taken together, we demonstrate that SYNPO is induced by IL-13 in vitro and in vivo, is a nonredundant regulator of epithelial cell barrier function and motility, and is likely involved in EoE pathogenesis.
Figures


Similar articles
-
Preferential Secretion of Thymic Stromal Lymphopoietin (TSLP) by Terminally Differentiated Esophageal Epithelial Cells: Relevance to Eosinophilic Esophagitis (EoE).PLoS One. 2016 Mar 18;11(3):e0150968. doi: 10.1371/journal.pone.0150968. eCollection 2016. PLoS One. 2016. PMID: 26992000 Free PMC article.
-
LRRC31 is induced by IL-13 and regulates kallikrein expression and barrier function in the esophageal epithelium.Mucosal Immunol. 2016 May;9(3):744-56. doi: 10.1038/mi.2015.98. Epub 2015 Oct 14. Mucosal Immunol. 2016. PMID: 26462420 Free PMC article.
-
IL-33 is induced in undifferentiated, non-dividing esophageal epithelial cells in eosinophilic esophagitis.Sci Rep. 2017 Dec 14;7(1):17563. doi: 10.1038/s41598-017-17541-5. Sci Rep. 2017. PMID: 29242581 Free PMC article.
-
Molecular basis and cellular mechanisms of eosinophilic esophagitis for the clinical practice.Expert Rev Gastroenterol Hepatol. 2019 Feb;13(2):99-117. doi: 10.1080/17474124.2019.1546120. Epub 2018 Nov 15. Expert Rev Gastroenterol Hepatol. 2019. PMID: 30791784 Review.
-
Eosinophils in Eosinophilic Esophagitis: The Road to Fibrostenosis is Paved With Good Intentions.Front Immunol. 2020 Dec 1;11:603295. doi: 10.3389/fimmu.2020.603295. eCollection 2020. Front Immunol. 2020. PMID: 33335531 Free PMC article. Review.
Cited by
-
Silence of IGFBP7 suppresses apoptosis and epithelial mesenchymal transformation of high glucose induced-podocytes.Exp Ther Med. 2018 Aug;16(2):1095-1102. doi: 10.3892/etm.2018.6298. Epub 2018 Jun 13. Exp Ther Med. 2018. PMID: 30112052 Free PMC article.
-
Genetic, Inflammatory, and Epithelial Cell Differentiation Factors Control Expression of Human Calpain-14.G3 (Bethesda). 2019 Mar 7;9(3):729-736. doi: 10.1534/g3.118.200901. G3 (Bethesda). 2019. PMID: 30626591 Free PMC article.
-
Mechanistic Insights into Eosinophilic Esophagitis: Therapies Targeting Pathophysiological Mechanisms.Cells. 2023 Oct 18;12(20):2473. doi: 10.3390/cells12202473. Cells. 2023. PMID: 37887317 Free PMC article. Review.
-
The occurrence and development mechanisms of esophageal stricture: state of the art review.J Transl Med. 2024 Jan 31;22(1):123. doi: 10.1186/s12967-024-04932-2. J Transl Med. 2024. PMID: 38297325 Free PMC article. Review.
-
Esophageal type 2 cytokine expression heterogeneity in eosinophilic esophagitis in a multisite cohort.J Allergy Clin Immunol. 2020 Jun;145(6):1629-1640.e4. doi: 10.1016/j.jaci.2020.01.051. Epub 2020 Mar 19. J Allergy Clin Immunol. 2020. PMID: 32197970 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

