Maximizing ecological and evolutionary insight in bisulfite sequencing data sets
- PMID: 29046582
- PMCID: PMC5656403
- DOI: 10.1038/s41559-017-0229-0
Maximizing ecological and evolutionary insight in bisulfite sequencing data sets
Abstract
Genome-scale bisulfite sequencing approaches have opened the door to ecological and evolutionary studies of DNA methylation in many organisms. These approaches can be powerful. However, they introduce new methodological and statistical considerations, some of which are particularly relevant to non-model systems. Here, we highlight how these considerations influence a study's power to link methylation variation with a predictor variable of interest. Relative to current practice, we argue that sample sizes will need to increase to provide robust insights. We also provide recommendations for overcoming common challenges and an R Shiny app to aid in study design.
Conflict of interest statement
We declare no conflict of interest.
Figures
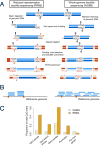
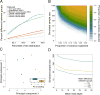
References
-
- Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 2011;13:97–109. - PubMed
-
- Jones P. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–92. - PubMed
-
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. - PubMed
-
- Seymour DK, Becker C. The causes and consequences of DNA methylome variation in plants. Curr. Opin. Plant Biol. 2017;36:56–63. - PubMed
-
- Verhoeven KJF, Jansen JJ, van Dijk PJ, Biere A. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 2010;185:1108–1118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

