Comparative Analysis of the Complete Plastomes of Apostasia wallichii and Neuwiedia singapureana (Apostasioideae) Reveals Different Evolutionary Dynamics of IR/SSC Boundary among Photosynthetic Orchids
- PMID: 29046685
- PMCID: PMC5632729
- DOI: 10.3389/fpls.2017.01713
Comparative Analysis of the Complete Plastomes of Apostasia wallichii and Neuwiedia singapureana (Apostasioideae) Reveals Different Evolutionary Dynamics of IR/SSC Boundary among Photosynthetic Orchids
Abstract
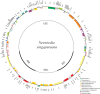
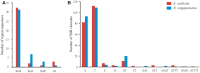
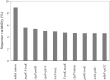
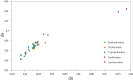
Apostasioideae, consists of only two genera, Apostasia and Neuwiedia, which are mainly distributed in Southeast Asia and northern Australia. The floral structure, taxonomy, biogeography, and genome variation of Apostasioideae have been intensively studied. However, detailed analyses of plastome composition and structure and comparisons with those of other orchid subfamilies have not yet been conducted. Here, the complete plastome sequences of Apostasia wallichii and Neuwiedia singapureana were sequenced and compared with 43 previously published photosynthetic orchid plastomes to characterize the plastome structure and evolution in the orchids. Unlike many orchid plastomes (e.g., Paphiopedilum and Vanilla), the plastomes of Apostasioideae contain a full set of 11 functional NADH dehydrogenase (ndh) genes. The distribution of repeat sequences and simple sequence repeat elements enhanced the view that the mutation rate of non-coding regions was higher than that of coding regions. The 10 loci-ndhA intron, matK-5'trnK, clpP-psbB, rps8-rpl14, trnT-trnL, 3'trnK-matK, clpP intron, psbK-trnK, trnS-psbC, and ndhF-rpl32-that had the highest degrees of sequence variability were identified as mutational hotspots for the Apostasia plastome. Furthermore, our results revealed that plastid genes exhibited a variable evolution rate within and among different orchid genus. Considering the diversified evolution of both coding and non-coding regions, we suggested that the plastome-wide evolution of orchid species was disproportional. Additionally, the sequences flanking the inverted repeat/small single copy (IR/SSC) junctions of photosynthetic orchid plastomes were categorized into three types according to the presence/absence of ndh genes. Different evolutionary dynamics for each of the three IR/SSC types of photosynthetic orchid plastomes were also proposed.
Keywords: Apostasioideae; IR expansion/contraction; hotspots; plastome; substitution rates.
Figures

Similar articles
-
Plastome Evolution and Phylogeny of Orchidaceae, With 24 New Sequences.Front Plant Sci. 2020 Feb 21;11:22. doi: 10.3389/fpls.2020.00022. eCollection 2020. Front Plant Sci. 2020. PMID: 32153600 Free PMC article.
-
The Complete Plastome Sequences of Four Orchid Species: Insights into the Evolution of the Orchidaceae and the Utility of Plastomic Mutational Hotspots.Front Plant Sci. 2017 May 3;8:715. doi: 10.3389/fpls.2017.00715. eCollection 2017. Front Plant Sci. 2017. PMID: 28515737 Free PMC article.
-
The chloroplast genome evolution of Venus slipper (Paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions.BMC Plant Biol. 2021 May 31;21(1):248. doi: 10.1186/s12870-021-03053-y. BMC Plant Biol. 2021. PMID: 34058997 Free PMC article.
-
Dynamic evolution of the plastome in the Elm family (Ulmaceae).Planta. 2022 Dec 17;257(1):14. doi: 10.1007/s00425-022-04045-4. Planta. 2022. PMID: 36526857 Review.
-
A comparative analysis of plastome evolution in autotrophic Piperales.Am J Bot. 2024 Mar;111(3):e16300. doi: 10.1002/ajb2.16300. Epub 2024 Mar 12. Am J Bot. 2024. PMID: 38469876 Review.
Cited by
-
Plastid phylogenomics of Pleurothallidinae (Orchidaceae): Conservative plastomes, new variable markers, and comparative analyses of plastid, nuclear, and mitochondrial data.PLoS One. 2021 Aug 27;16(8):e0256126. doi: 10.1371/journal.pone.0256126. eCollection 2021. PLoS One. 2021. PMID: 34449781 Free PMC article.
-
The complete plastid genomes of Ophrys iricolor and O. sphegodes (Orchidaceae) and comparative analyses with other orchids.PLoS One. 2018 Sep 18;13(9):e0204174. doi: 10.1371/journal.pone.0204174. eCollection 2018. PLoS One. 2018. PMID: 30226857 Free PMC article.
-
Mutational Biases and GC-Biased Gene Conversion Affect GC Content in the Plastomes of Dendrobium Genus.Int J Mol Sci. 2017 Nov 2;18(11):2307. doi: 10.3390/ijms18112307. Int J Mol Sci. 2017. PMID: 29099062 Free PMC article.
-
The complete chloroplast genome of Cyrtosia nana (Rolfe ex Downie) Garay and its phylogenetic analysis.Mitochondrial DNA B Resour. 2025 Jul 9;10(8):683-686. doi: 10.1080/23802359.2025.2528572. eCollection 2025. Mitochondrial DNA B Resour. 2025. PMID: 40642020 Free PMC article.
-
A novel seed dispersal mode of Apostasia nipponica could provide some clues to the early evolution of the seed dispersal system in Orchidaceae.Evol Lett. 2020 Aug 2;4(5):457-464. doi: 10.1002/evl3.188. eCollection 2020 Oct. Evol Lett. 2020. PMID: 33014421 Free PMC article.
References
-
- Ahmed I., Matthews P. J., Biggs P. J., Naeem M., Mclenachan P. A., Lockhart P. J. (2013). Identification of chloroplast genome loci suitable for high-resolution phylogeographic studies of Colocasia esculenta (L.) Schott (Araceae) and closely related taxa. Mol. Ecol. Resour. 13 929–937. 10.1111/1755-0998.12128 - DOI - PubMed
-
- Chang C. C., Lin H. C., Lin I. P., Chow T. Y., Chen H. H., Chen W. H., et al. (2006). The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol. Biol. Evol. 23 279–291. 10.1093/molbev/msj029 - DOI - PubMed
-
- Chase M. W., Cameron K. M., Barrett R. L., Freudenstein J. V. (2003). “DNA data and Orchidaceae systematics: a new phylogenetic classification,” in Orchid Conservation, eds Dixon K. W., Kell S. P., Barrett R. L., Cribb P. J. (Kota Kinabalu: Natural History Press; ), 69–89.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

