Increased lipid production by heterologous expression of AtWRI1 transcription factor in Nannochloropsis salina
- PMID: 29046718
- PMCID: PMC5635583
- DOI: 10.1186/s13068-017-0919-5
Increased lipid production by heterologous expression of AtWRI1 transcription factor in Nannochloropsis salina
Abstract
Background: Genetic engineering of microalgae is necessary to produce economically feasible strains for biofuel production. Current efforts are focused on the manipulation of individual metabolic genes, but the outcomes are not sufficiently stable and/or efficient for large-scale production of biofuels and other materials. Transcription factors (TFs) are emerging as good alternatives for engineering of microalgae, not only to increase production of biomaterials but to enhance stress tolerance. Here, we investigated an AP2 type TF Wrinkled1 in Arabidopsis (AtWRI1) known as a key regulator of lipid biosynthesis in plants, and applied it to industrial microalgae, Nannochloropsis salina.
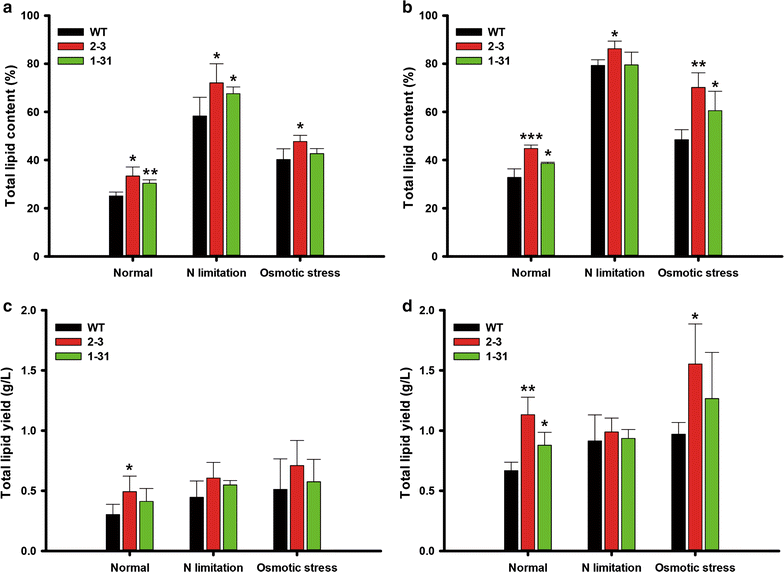
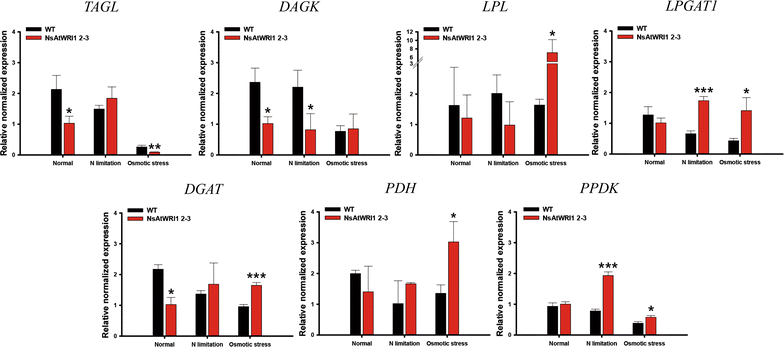
Results: We expressed AtWRI1 TF heterologously in N. salina, named NsAtWRI1, in an effort to re-enact its key regulatory function of lipid accumulation. Stable integration AtWRI1 was confirmed by RESDA PCR, and its expression was confirmed by Western blotting using the FLAG tag. Characterizations of transformants revealed that the neutral and total lipid contents were greater in NsAtWRI1 transformants than in WT under both normal and stress conditions from day 8. Especially, total lipid contents were 36.5 and 44.7% higher in NsAtWRI1 2-3 than in WT under normal and osmotic stress condition, respectively. FAME contents of NsAtWRI1 2-3 were also increased compared to WT. As a result, FAME yield of NsAtWRI1 2-3 was increased to 768 mg/L/day, which was 64% higher than that of WT under the normal condition. We identified candidates of AtWRI1-regulated genes by searching for the presence of the AW-box in promoter regions, among which lipid metabolic genes were further analyzed by qRT-PCR. Overall, qRT-PCR results on day 1 indicated that AtWRI1 down-regulated TAGL and DAGK, and up-regulated PPDK, LPL, LPGAT1, and PDH, resulting in enhanced lipid production in NsAtWRI1 transformants from early growth phase.
Conclusion: AtWRI1 TF regulated several genes involved in lipid synthesis in N. salina, resulting in enhancement of neutral lipid and FAME production. These findings suggest that heterologous expression of AtWRI1 TF can be utilized for efficient biofuel production in industrial microalgae.
Keywords: Biofuels; Microalgae; Nannochloropsis salina; TF engineering; Transcription factor; Wrinkled1.
Figures



Similar articles
-
Heterologous synthesis of chlorophyll b in Nannochloropsis salina enhances growth and lipid production by increasing photosynthetic efficiency.Biotechnol Biofuels. 2019 May 14;12:122. doi: 10.1186/s13068-019-1462-3. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31114631 Free PMC article.
-
Effects of overexpression of a bHLH transcription factor on biomass and lipid production in Nannochloropsis salina.Biotechnol Biofuels. 2015 Dec 1;8:200. doi: 10.1186/s13068-015-0386-9. eCollection 2015. Biotechnol Biofuels. 2015. PMID: 26628914 Free PMC article.
-
Enhancement of biomass and lipid productivity by overexpression of a bZIP transcription factor in Nannochloropsis salina.Biotechnol Bioeng. 2018 Feb;115(2):331-340. doi: 10.1002/bit.26465. Epub 2017 Nov 3. Biotechnol Bioeng. 2018. PMID: 28976541
-
Biomass and lipid induction strategies in microalgae for biofuel production and other applications.Microb Cell Fact. 2019 Oct 21;18(1):178. doi: 10.1186/s12934-019-1228-4. Microb Cell Fact. 2019. PMID: 31638987 Free PMC article. Review.
-
Enhancement of lipid accumulation in microalgae by metabolic engineering.Biochim Biophys Acta Mol Cell Biol Lipids. 2019 Apr;1864(4):552-566. doi: 10.1016/j.bbalip.2018.10.004. Epub 2018 Oct 8. Biochim Biophys Acta Mol Cell Biol Lipids. 2019. PMID: 30308323 Review.
Cited by
-
Heterologous synthesis of chlorophyll b in Nannochloropsis salina enhances growth and lipid production by increasing photosynthetic efficiency.Biotechnol Biofuels. 2019 May 14;12:122. doi: 10.1186/s13068-019-1462-3. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31114631 Free PMC article.
-
Engineering the Metabolic Landscape of Microorganisms for Lignocellulosic Conversion.Microorganisms. 2023 Aug 31;11(9):2197. doi: 10.3390/microorganisms11092197. Microorganisms. 2023. PMID: 37764041 Free PMC article. Review.
-
NgAP2a Targets KCS Gene to Promote Lipid Accumulation in Nannochloropsis gaditana.Int J Mol Sci. 2024 Sep 25;25(19):10305. doi: 10.3390/ijms251910305. Int J Mol Sci. 2024. PMID: 39408634 Free PMC article.
-
Microalgae biofuels: illuminating the path to a sustainable future amidst challenges and opportunities.Biotechnol Biofuels Bioprod. 2024 Jan 23;17(1):10. doi: 10.1186/s13068-024-02461-0. Biotechnol Biofuels Bioprod. 2024. PMID: 38254224 Free PMC article. Review.
-
Enhancing fatty acid and omega-3 production in Schizochytrium sp. using developed safe-harboring expression system.J Biol Eng. 2024 Oct 10;18(1):56. doi: 10.1186/s13036-024-00447-y. J Biol Eng. 2024. PMID: 39390586 Free PMC article.
References
-
- Williams PJlB, Laurens LML. Microalgae as biodiesel & biomass feedstocks: Review & analysis of the biochemistry, energetics & economics. Energy Environ Sci. 2010;3:554. doi: 10.1039/b924978h. - DOI
-
- Mata TM, Martins AA, Caetano NS. Microalgae for biodiesel production and other applications: a review. Renew Sust Energ Rev. 2010;14:217–232. doi: 10.1016/j.rser.2009.07.020. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

