Emergence and loss of spliceosomal twin introns
- PMID: 29046814
- PMCID: PMC5639578
- DOI: 10.1186/s40694-017-0037-y
Emergence and loss of spliceosomal twin introns
Abstract
Background: In the primary transcript of nuclear genes, coding sequences-exons-usually alternate with non-coding sequences-introns. In the evolution of spliceosomal intron-exon structure, extant intron positions can be abandoned and new intron positions can be occupied. Spliceosomal twin introns ("stwintrons") are unconventional intervening sequences where a standard "internal" intron interrupts a canonical splicing motif of a second, "external" intron. The availability of genome sequences of more than a thousand species of fungi provides a unique opportunity to study spliceosomal intron evolution throughout a whole kingdom by means of molecular phylogenetics.
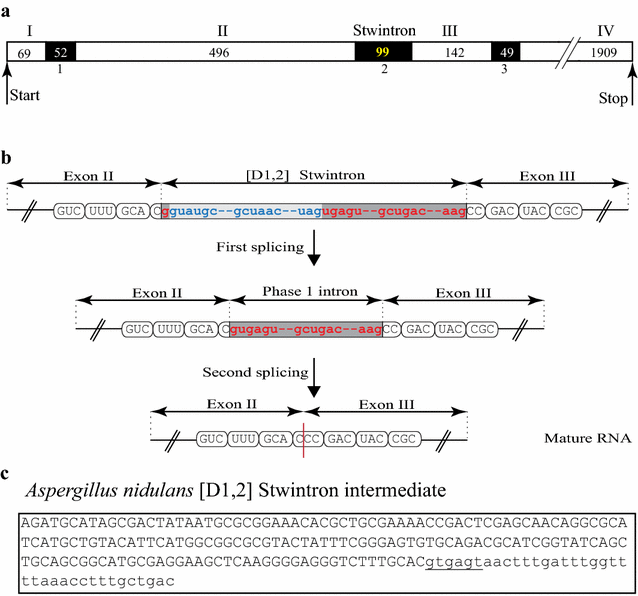
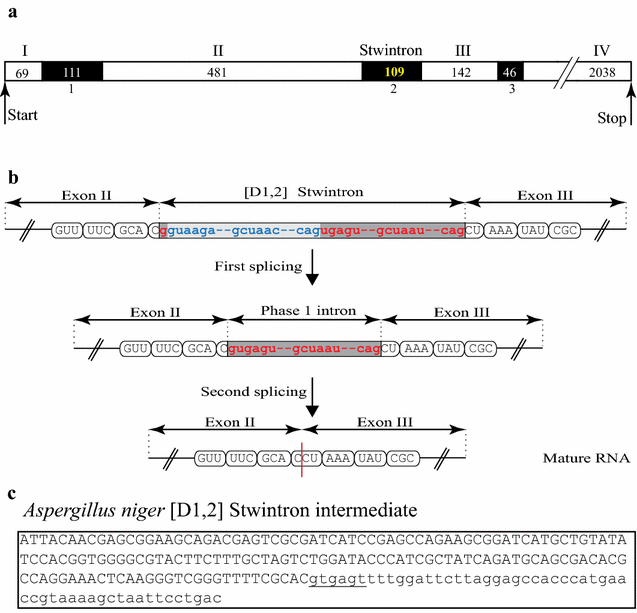
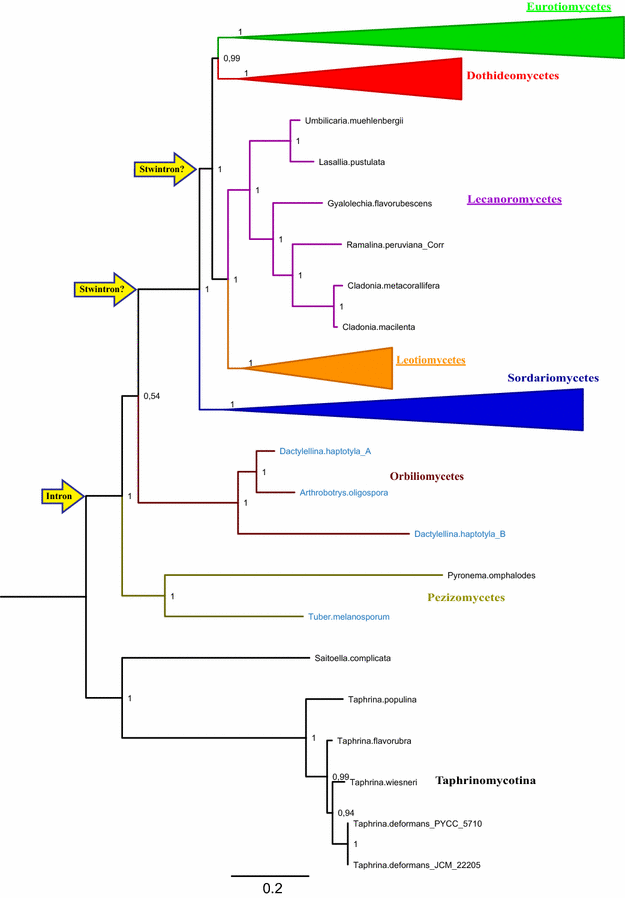
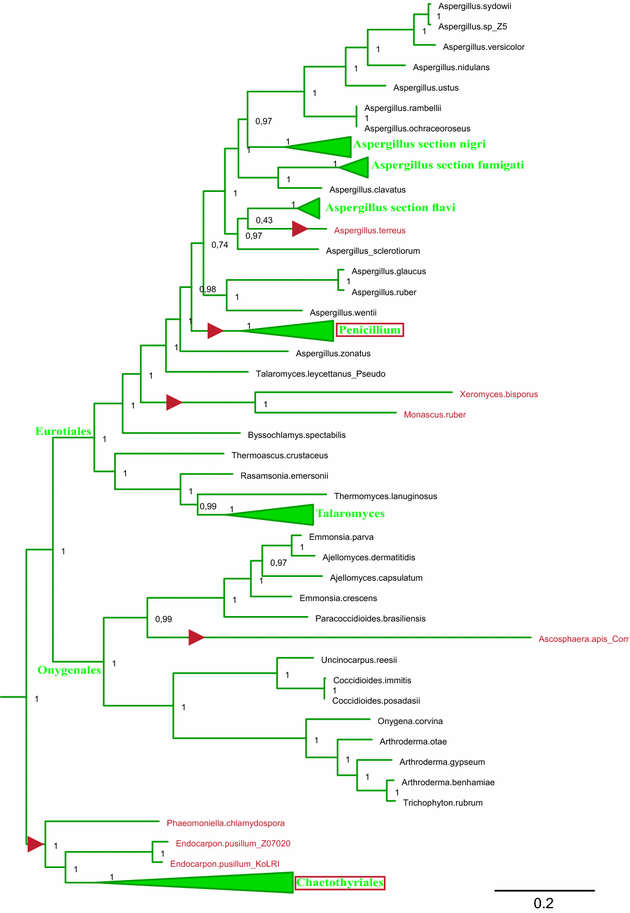
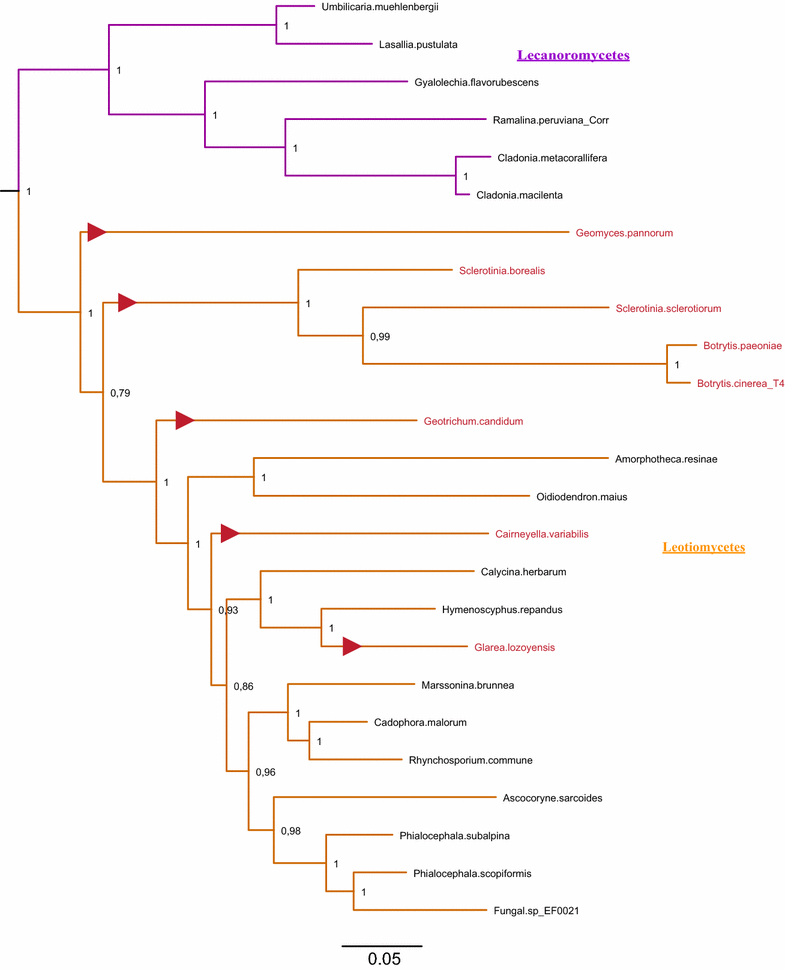
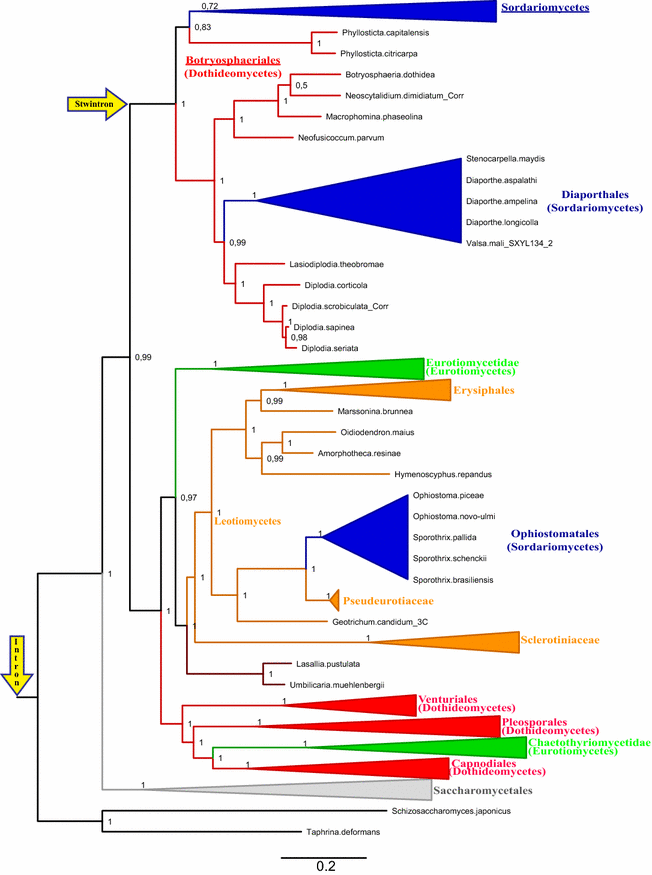
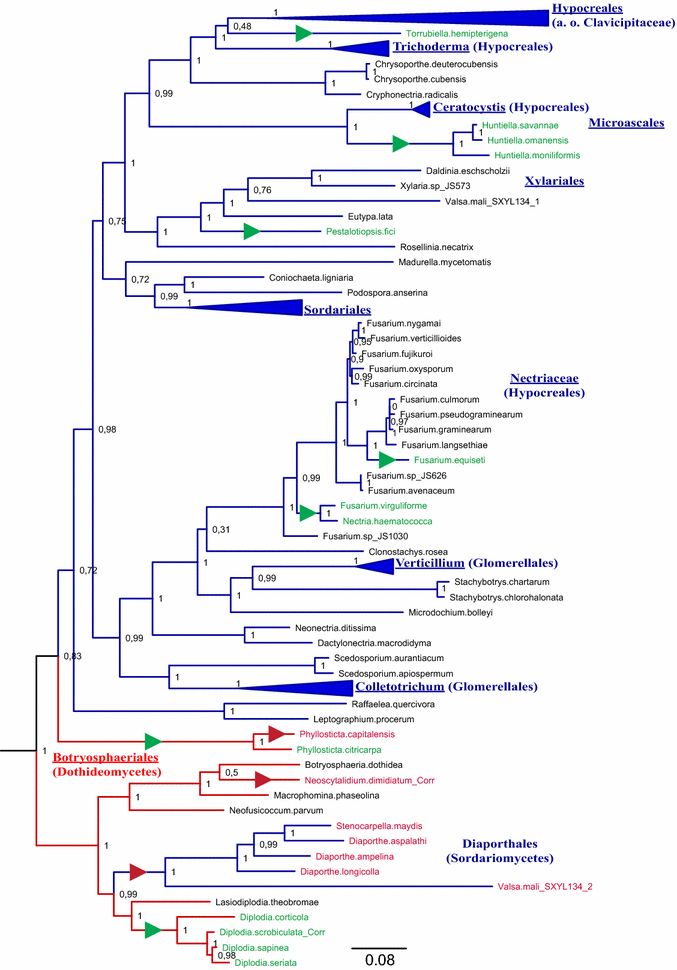
Results: A new stwintron was encountered in Aspergillus nidulans and Aspergillus niger. It is present across three classes of Leotiomyceta in the transcript of a well-conserved gene encoding a putative lipase (lipS). It occupies the same position as a standard intron in the orthologue gene in species of the early divergent classes of the Pezizomycetes and the Orbiliomycetes, suggesting that an internal intron has appeared within a pre-extant intron. On the other hand, the stwintron has been lost from certain taxa in Leotiomycetes and Eurotiomycetes at several occasions, most likely by a mechanism involving reverse transcription and homologous recombination. Another ancient stwintron present across whole Pezizomycotina orders-in the transcript of the bifunctional biotin biosynthesis gene bioDA-occurs at the same position as a standard intron in many species of non-Dikarya. Nevertheless, also the bioDA stwintron has disappeared from certain lineages within the taxa where it occurs, i.e., Sordariomycetes and Botryosphaeriales. Intriguingly, only the internal intron was lost from the Sordariomycetes bioDA stwintron at all but one occasion, leaving a standard intron in the same position, while where the putative lipase stwintron was lost, no intronic sequences remain.
Conclusions: Molecular phylogeny of the peptide product was used to monitor the existence and fate of a stwintron in the transcripts of two neatly defined fungal genes, encoding well conserved proteins. Both defining events-stwintron emergence and loss-can be explained with extant models for intron insertion and loss. We thus demonstrate that stwintrons can serve as model systems to study spliceosomal intron evolution.
Keywords: Aspergillus nidulans; Intron gain; Intron loss; Molecular phylogenetics; Pezizomycotina; Spliceosomal intron evolution; Spliceosomal twin introns.
Figures
Similar articles
-
A spliceosomal twin intron (stwintron) participates in both exon skipping and evolutionary exon loss.Sci Rep. 2019 Jul 9;9(1):9940. doi: 10.1038/s41598-019-46435-x. Sci Rep. 2019. PMID: 31289343 Free PMC article.
-
Propagation of [D1,2]-type spliceosomal twin introns (stwintrons) in Hypoxylaceae and Xylariaceae fungi.Microbiol Spectr. 2025 Aug 8:e0292624. doi: 10.1128/spectrum.02926-24. Online ahead of print. Microbiol Spectr. 2025. PMID: 40778755
-
A mechanism for a single nucleotide intron shift.Nucleic Acids Res. 2017 Sep 6;45(15):9085-9092. doi: 10.1093/nar/gkx520. Nucleic Acids Res. 2017. PMID: 28595329 Free PMC article.
-
Models of spliceosomal intron proliferation in the face of widespread ectopic expression.Gene. 2006 Feb 1;366(2):201-8. doi: 10.1016/j.gene.2005.09.004. Epub 2005 Nov 8. Gene. 2006. PMID: 16288838 Review.
-
Analysis of evolution of exon-intron structure of eukaryotic genes.Brief Bioinform. 2005 Jun;6(2):118-34. doi: 10.1093/bib/6.2.118. Brief Bioinform. 2005. PMID: 15975222 Review.
Cited by
-
Internally Symmetrical Stwintrons and Related Canonical Introns in Hypoxylaceae Species.J Fungi (Basel). 2021 Aug 29;7(9):710. doi: 10.3390/jof7090710. J Fungi (Basel). 2021. PMID: 34575748 Free PMC article.
-
Advances in understanding the evolution of fungal genome architecture.F1000Res. 2020 Jul 27;9:F1000 Faculty Rev-776. doi: 10.12688/f1000research.25424.1. eCollection 2020. F1000Res. 2020. PMID: 32765832 Free PMC article. Review.
-
Unique and Repeated Stwintrons (Spliceosomal Twin Introns) in the Hypoxylaceae.J Fungi (Basel). 2022 Apr 13;8(4):397. doi: 10.3390/jof8040397. J Fungi (Basel). 2022. PMID: 35448628 Free PMC article.
-
A spliceosomal twin intron (stwintron) participates in both exon skipping and evolutionary exon loss.Sci Rep. 2019 Jul 9;9(1):9940. doi: 10.1038/s41598-019-46435-x. Sci Rep. 2019. PMID: 31289343 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

