GSK-J4-Mediated Transcriptomic Alterations in Differentiating Embryoid Bodies
- PMID: 29047260
- PMCID: PMC5682251
- DOI: 10.14348/molcells.2017.0069
GSK-J4-Mediated Transcriptomic Alterations in Differentiating Embryoid Bodies
Abstract
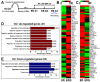
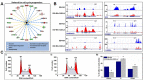
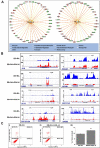
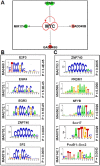
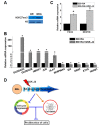
Histone-modifying enzymes are key players in the field of cellular differentiation. Here, we used GSK-J4 to profile important target genes that are responsible for neural differentiation. Embryoid bodies were treated with retinoic acid (10 μM) to induce neural differentiation in the presence or absence of GSK-J4. To profile GSKJ4-target genes, we performed RNA sequencing for both normal and demethylase-inhibited cells. A total of 47 and 58 genes were up- and down-regulated, respectively, after GSK-J4 exposure at a log2-fold-change cut-off value of 1.2 (p-value < 0.05). Functional annotations of all of the differentially expressed genes revealed that a significant number of genes were associated with the suppression of cellular proliferation, cell cycle progression and induction of cell death. We also identified an enrichment of potent motifs in selected genes that were differentially expressed. Additionally, we listed upstream transcriptional regulators of all of the differentially expressed genes. Our data indicate that GSK-J4 affects cellular biology by inhibiting cellular proliferation through cell cycle suppression and induction of cell death. These findings will expand the current understanding of the biology of histone-modifying enzymes, thereby promoting further investigations to elucidate the underlying mechanisms.
Keywords: RNA sequencing; cell cycle progression; gene expression; histone demethylase enzyme.
Figures


Similar articles
-
Therapeutic potential of GSK-J4, a histone demethylase KDM6B/JMJD3 inhibitor, for acute myeloid leukemia.J Cancer Res Clin Oncol. 2018 Jun;144(6):1065-1077. doi: 10.1007/s00432-018-2631-7. Epub 2018 Mar 28. J Cancer Res Clin Oncol. 2018. PMID: 29594337 Free PMC article.
-
The antischistosomal potential of GSK-J4, an H3K27 demethylase inhibitor: insights from molecular modeling, transcriptomics and in vitro assays.Parasit Vectors. 2020 Mar 17;13(1):140. doi: 10.1186/s13071-020-4000-z. Parasit Vectors. 2020. PMID: 32178714 Free PMC article.
-
Oncogenic KRAS Sensitizes Lung Adenocarcinoma to GSK-J4-Induced Metabolic and Oxidative Stress.Cancer Res. 2019 Nov 15;79(22):5849-5859. doi: 10.1158/0008-5472.CAN-18-3511. Epub 2019 Sep 10. Cancer Res. 2019. PMID: 31506334
-
Gene expression signatures after ethanol exposure in differentiating embryoid bodies.Toxicol In Vitro. 2018 Feb;46:66-76. doi: 10.1016/j.tiv.2017.10.004. Epub 2017 Oct 3. Toxicol In Vitro. 2018. PMID: 28986285
-
GSK-J4: An H3K27 histone demethylase inhibitor, as a potential anti-cancer agent.Int J Cancer. 2023 Sep 15;153(6):1130-1138. doi: 10.1002/ijc.34559. Epub 2023 May 10. Int J Cancer. 2023. PMID: 37165737 Review.
Cited by
-
Integrated Omic Analyses Identify Pathways and Transcriptomic Regulators Associated With Chemical Alterations of In Vitro Neural Network Formation.Toxicol Sci. 2022 Feb 28;186(1):118-133. doi: 10.1093/toxsci/kfab151. Toxicol Sci. 2022. PMID: 34927697 Free PMC article.
-
Chromatin-modifying drugs and metabolites in cell fate control.Cell Prolif. 2020 Nov;53(11):e12898. doi: 10.1111/cpr.12898. Epub 2020 Sep 26. Cell Prolif. 2020. PMID: 32979011 Free PMC article. Review.
-
Epigenetic reprogramming of H3K27me3 and DNA methylation during leaf-to-callus transition in peach.Hortic Res. 2022 Jun 3;9:uhac132. doi: 10.1093/hr/uhac132. eCollection 2022. Hortic Res. 2022. PMID: 35937864 Free PMC article.
References
-
- Chen C., Chang Y.C., Lan M.S., Breslin M. Leptin stimulates ovarian cancer cell growth and inhibits apoptosis by increasing cyclin D1 and Mcl-1 expression via the activation of the MEK/ERK1/2 and PI3K/Akt signaling pathways. Int J Oncol. 2013;42:1113–1119. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

