Long-read sequencing of the coffee bean transcriptome reveals the diversity of full-length transcripts
- PMID: 29048540
- PMCID: PMC5737654
- DOI: 10.1093/gigascience/gix086
Long-read sequencing of the coffee bean transcriptome reveals the diversity of full-length transcripts
Abstract
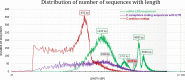
Polyploidization contributes to the complexity of gene expression, resulting in numerous related but different transcripts. This study explored the transcriptome diversity and complexity of the tetraploid Arabica coffee (Coffea arabica) bean. Long-read sequencing (LRS) by Pacbio Isoform sequencing (Iso-seq) was used to obtain full-length transcripts without the difficulty and uncertainty of assembly required for reads from short-read technologies. The tetraploid transcriptome was annotated and compared with data from the sub-genome progenitors. Caffeine and sucrose genes were targeted for case analysis. An isoform-level tetraploid coffee bean reference transcriptome with 95 995 distinct transcripts (average 3236 bp) was obtained. A total of 88 715 sequences (92.42%) were annotated with BLASTx against NCBI non-redundant plant proteins, including 34 719 high-quality annotations. Further BLASTn analysis against NCBI non-redundant nucleotide sequences, Coffea canephora coding sequences with UTR, C. arabica ESTs, and Rfam resulted in 1213 sequences without hits, were potential novel genes in coffee. Longer UTRs were captured, especially in the 5΄UTRs, facilitating the identification of upstream open reading frames. The LRS also revealed more and longer transcript variants in key caffeine and sucrose metabolism genes from this polyploid genome. Long sequences (>10 kilo base) were poorly annotated. LRS technology shows the limitation of previous studies. It provides an important tool to produce a reference transcriptome including more of the diversity of full-length transcripts to help understand the biology and support the genetic improvement of polyploid species such as coffee.
Keywords: UTR; coffee; full-length cDNA; isoform; long sequences; polyploid; transcriptome.
© The Authors 2017. Published by Oxford University Press.
Figures




References
-
- Yoo M, Liu X, Pires JC et al. Nonadditive gene expression in polyploids. Annu Rev Genet 2014;48(1):485–517. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

