Linkage of A-to-I RNA Editing in Metazoans and the Impact on Genome Evolution
- PMID: 29048557
- PMCID: PMC5850729
- DOI: 10.1093/molbev/msx274
Linkage of A-to-I RNA Editing in Metazoans and the Impact on Genome Evolution
Abstract
The adenosine-to-inosine (A-to-I) RNA editomes have been systematically characterized in various metazoan species, and many editing sites were found in clusters. However, it remains unclear whether the clustered editing sites tend to be linked in the same RNA molecules or not. By adopting a method originally designed to detect linkage disequilibrium of DNA mutations, we examined the editomes of ten metazoan species and detected extensive linkage of editing in Drosophila and cephalopods. The prevalent linkages of editing in these two clades, many of which are conserved between closely related species and might be associated with the adaptive proteomic recoding, are maintained by natural selection at the cost of genome evolution. Nevertheless, in worms and humans, we only detected modest proportions of linked editing events, the majority of which were not conserved. Furthermore, the linkage of editing in coding regions of worms and humans might be overall deleterious, which drives the evolution of DNA sites to escape promiscuous editing. Altogether, our results suggest that the linkage landscape of A-to-I editing has evolved during metazoan evolution. This present study also suggests that linkage of editing should be considered in elucidating the functional consequences of RNA editing.
Keywords: Drosophila; RNA editing; adaptive evolution; cephalopods; humans; linkage; mice; worms.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
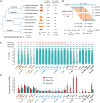
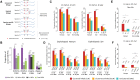
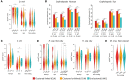



Similar articles
-
Conserved A-to-I RNA editing with non-conserved recoding expands the candidates of functional editing sites.Fly (Austin). 2024 Dec;18(1):2367359. doi: 10.1080/19336934.2024.2367359. Epub 2024 Jun 18. Fly (Austin). 2024. PMID: 38889318 Free PMC article.
-
The evolution and adaptation of A-to-I RNA editing.PLoS Genet. 2017 Nov 28;13(11):e1007064. doi: 10.1371/journal.pgen.1007064. eCollection 2017 Nov. PLoS Genet. 2017. PMID: 29182635 Free PMC article. Review.
-
Evolutionary Origins and Adaptive Significance of A-to-I RNA Editing in Animals and Fungi.Bioessays. 2025 May;47(5):e202400220. doi: 10.1002/bies.202400220. Epub 2025 Feb 21. Bioessays. 2025. PMID: 39981820 Review.
-
New comparative genomic evidence supporting the proteomic diversification role of A-to-I RNA editing in insects.Mol Genet Genomics. 2024 Apr 20;299(1):46. doi: 10.1007/s00438-024-02141-6. Mol Genet Genomics. 2024. PMID: 38642133
-
Learning from the Codon Table: Convergent Recoding Provides Novel Understanding on the Evolution of A-to-I RNA Editing.J Mol Evol. 2024 Aug;92(4):488-504. doi: 10.1007/s00239-024-10190-z. Epub 2024 Jul 16. J Mol Evol. 2024. PMID: 39012510
Cited by
-
A hierarchy in clusters of cephalopod mRNA editing sites.Sci Rep. 2022 Mar 2;12(1):3447. doi: 10.1038/s41598-022-07460-5. Sci Rep. 2022. PMID: 35236910 Free PMC article.
-
Illuminating spatial A-to-I RNA editing signatures within the Drosophila brain.Proc Natl Acad Sci U S A. 2019 Feb 5;116(6):2318-2327. doi: 10.1073/pnas.1811768116. Epub 2019 Jan 18. Proc Natl Acad Sci U S A. 2019. PMID: 30659150 Free PMC article.
-
ADAR Family Proteins: A Structural Review.Curr Issues Mol Biol. 2024 Apr 26;46(5):3919-3945. doi: 10.3390/cimb46050243. Curr Issues Mol Biol. 2024. PMID: 38785511 Free PMC article. Review.
-
6mA-Sniper: Quantifying 6mA sites in eukaryotes at single-nucleotide resolution.Sci Adv. 2023 Oct 20;9(42):eadh7912. doi: 10.1126/sciadv.adh7912. Epub 2023 Oct 20. Sci Adv. 2023. PMID: 37862411 Free PMC article.
-
Accurate identification of RNA editing sites from primitive sequence with deep neural networks.Sci Rep. 2018 Apr 16;8(1):6005. doi: 10.1038/s41598-018-24298-y. Sci Rep. 2018. PMID: 29662087 Free PMC article.
References
-
- Barrett JC, Fry B, Maller J, Daly MJ.. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2): 263–265.http://dx.doi.org/10.1093/bioinformatics/bth457 - DOI - PubMed
-
- Bass BL. 2002. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 71:817–846.http://dx.doi.org/10.1146/annurev.biochem.71.110601.135501 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

