Linear discriminant analysis reveals differences in root architecture in wheat seedlings related to nitrogen uptake efficiency
- PMID: 29048563
- PMCID: PMC5853436
- DOI: 10.1093/jxb/erx300
Linear discriminant analysis reveals differences in root architecture in wheat seedlings related to nitrogen uptake efficiency
Abstract
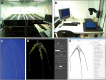
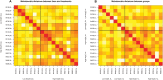
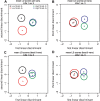
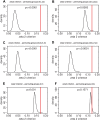
Root architecture impacts water and nutrient uptake efficiency. Identifying exactly which root architectural properties influence these agronomic traits can prove challenging. In this paper, approximately 300 wheat (Triticum aestivum) plants were divided into four groups using two binary classifications, high versus low nitrogen uptake efficiency (NUpE), and high versus low nitrate in the growth medium. The root system architecture for each wheat plant was captured using 16 quantitative variables. The multivariate analysis tool, linear discriminant analysis, was used to construct composite variables, each a linear combination of the original variables, such that the score of the plants on the new variables showed the maximum between-group variability. The results show that the distribution of root-system architecture traits differs between low- and high-NUpE plants and, less strongly, between low-NUpE plants grown on low versus high nitrate media.
Keywords: Linear discriminant analysis; Mahalanobis distance; Triticum aestivum; Watkins lines; nitrogen uptake efficiency; plant phenotyping; root system architecture; wheat root biology.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures





References
-
- Bullitt E, Liu A, Aylward SR, Coffey C, Stone J, Mukherji SK, Muller KE, Pizer SM. 1999. Registration of 3D cerebral vessels with 2D digital angiograms: clinical evaluation. Academic Radiology 6, 539–546. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

