The yeast 2-μm plasmid Raf protein contributes to plasmid inheritance by stabilizing the Rep1 and Rep2 partitioning proteins
- PMID: 29048592
- PMCID: PMC5737570
- DOI: 10.1093/nar/gkx703
The yeast 2-μm plasmid Raf protein contributes to plasmid inheritance by stabilizing the Rep1 and Rep2 partitioning proteins
Abstract
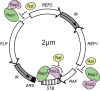
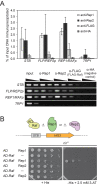
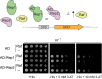
The yeast 2-μm plasmid is a remarkable genetic parasite, managing efficient maintenance at high-copy number with minimal impact on the host. Equal partitioning of the plasmid upon host cell division requires plasmid proteins Rep1 and Rep2 and the plasmid STB locus. The Rep proteins and the plasmid-encoded Raf protein also regulate plasmid gene transcription. In this study, protein interaction assays, sequence analyses and mutational approaches were used to identify domains and residues in Rep2 and Raf required for association with Rep1 and Rep2 and to delineate the Rep2 DNA-binding domain. Rep2 and Raf displayed similarities in interactions with Rep1 and Rep2, in having Rep1 promote their STB association in vivo, and in stabilizing Rep protein levels. Rep2 mutants impaired for self-association were competent for transcriptional repression while those deficient for Rep1 association were not. Surprisingly, Rep2 mutants impaired for either Rep1 interaction or self-association were able to maintain efficient plasmid inheritance provided Raf was present and competent for Rep protein interaction. Our findings provide insight into the Rep protein complexes required for partitioning and transcriptional repression, and suggest that in addition to its transcriptional function, Raf stabilization of Rep partitioning proteins contributes to the remarkable persistence of the 2-μm plasmid.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






References
-
- Chan K.M., Liu Y.T., Ma C.H., Jayaram M., Sau S.. The 2 micron plasmid of Saccharomyces cerevisiae: a miniaturized selfish genome with optimized functional competence. Plasmid. 2013; 70:2–17. - PubMed
-
- Blaisonneau J., Sor F., Cheret G., Yarrow D., Fukuhara H.. A circular plasmid from the yeast Torulaspora delbrueckii. Plasmid. 1997; 38:202–209. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

