Host blood RNA signatures predict the outcome of tuberculosis treatment
- PMID: 29050771
- PMCID: PMC5658513
- DOI: 10.1016/j.tube.2017.08.004
Host blood RNA signatures predict the outcome of tuberculosis treatment
Abstract
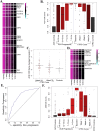
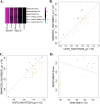
Biomarkers for tuberculosis treatment outcome will assist in guiding individualized treatment and evaluation of new therapies. To identify candidate biomarkers, RNA sequencing of whole blood from a well-characterized TB treatment cohort was performed. Application of a validated transcriptional correlate of risk for TB revealed symmetry in host gene expression during progression from latent TB infection to active TB disease and resolution of disease during treatment, including return to control levels after drug therapy. The symmetry was also seen in a TB disease signature, constructed from the TB treatment cohort, that also functioned as a strong correlate of risk. Both signatures identified patients at risk of treatment failure 1-4 weeks after start of therapy. Further mining of the transcriptomes revealed an association between treatment failure and suppressed expression of mitochondrial genes before treatment initiation, leading to development of a novel baseline (pre-treatment) signature of treatment failure. These novel host responses to TB treatment were integrated into a five-gene real-time PCR-based signature that captures the clinically relevant responses to TB treatment and provides a convenient platform for stratifying patients according to their risk of treatment failure. Furthermore, this 5-gene signature is shown to correlate with the pulmonary inflammatory state (as measured by PET-CT) and can complement sputum-based Gene Xpert for patient stratification, providing a rapid and accurate alternative to current methods.
Keywords: Biomarkers; Host response; Mitochondria; Transcriptome; Tuberculosis treatment.
Copyright © 2017 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

