Mechanisms of Phosphorus Acquisition and Lipid Class Remodeling under P Limitation in a Marine Microalga
- PMID: 29051196
- PMCID: PMC5717724
- DOI: 10.1104/pp.17.00621
Mechanisms of Phosphorus Acquisition and Lipid Class Remodeling under P Limitation in a Marine Microalga
Abstract
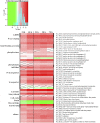
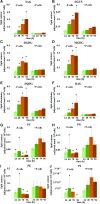
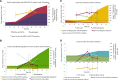
Molecular mechanisms of phosphorus (P) limitation are of great interest for understanding algal production in aquatic ecosystems. Previous studies point to P limitation-induced changes in lipid composition. As, in microalgae, the molecular mechanisms of this specific P stress adaptation remain unresolved, we reveal a detailed phospholipid-recycling scheme in Nannochloropsis oceanica and describe important P acquisition genes based on highly corresponding transcriptome and lipidome data. Initial responses to P limitation showed increased expression of genes involved in P uptake and an expansion of the P substrate spectrum based on purple acid phosphatases. Increase in P trafficking displayed a rearrangement between compartments by supplying P to the chloroplast and carbon to the cytosol for lipid synthesis. We propose a novel phospholipid-recycling scheme for algae that leads to the rapid reduction of phospholipids and synthesis of the P-free lipid classes. P mobilization through membrane lipid degradation is mediated mainly by two glycerophosphoryldiester phosphodiesterases and three patatin-like phospholipases A on the transcriptome level. To compensate for low phospholipids in exponential growth, N. oceanica synthesized sulfoquinovosyldiacylglycerol and diacylglyceroltrimethylhomoserine. In this study, it was shown that an N. oceanica strain has a unique repertoire of genes that facilitate P acquisition and the degradation of phospholipids compared with other stramenopiles. The novel phospholipid-recycling scheme opens new avenues for metabolic engineering of lipid composition in algae.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures


References
-
- Abdullahi AS, Underwood GJ, Gretz MR (2006) Extracellular matrix assembly in diatoms (Bacillariophyceae). J Phycol 42: 363–378
-
- Bates PD, Browse J (2011) The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J 68: 387–399 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

