D4 dopamine receptor high-resolution structures enable the discovery of selective agonists
- PMID: 29051383
- PMCID: PMC5856174
- DOI: 10.1126/science.aan5468
D4 dopamine receptor high-resolution structures enable the discovery of selective agonists
Abstract
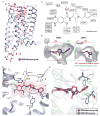
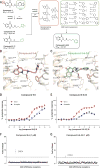
Dopamine receptors are implicated in the pathogenesis and treatment of nearly every neuropsychiatric disorder. Although thousands of drugs interact with these receptors, our molecular understanding of dopaminergic drug selectivity and design remains clouded. To illuminate dopamine receptor structure, function, and ligand recognition, we determined crystal structures of the D4 dopamine receptor in its inactive state bound to the antipsychotic drug nemonapride, with resolutions up to 1.95 angstroms. These structures suggest a mechanism for the control of constitutive signaling, and their unusually high resolution enabled a structure-based campaign for new agonists of the D4 dopamine receptor. The ability to efficiently exploit structure for specific probe discovery-rapidly moving from elucidating receptor structure to discovering previously unrecognized, selective agonists-testifies to the power of structure-based approaches.
Copyright © 2017 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

