Genomic landscape associated with potential response to anti-CTLA-4 treatment in cancers
- PMID: 29051489
- PMCID: PMC5648801
- DOI: 10.1038/s41467-017-01018-0
Genomic landscape associated with potential response to anti-CTLA-4 treatment in cancers
Abstract
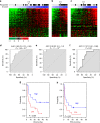
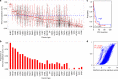
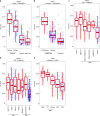
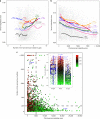
Immunotherapy has emerged as a promising anti-cancer treatment, however, little is known about the genetic characteristics that dictate response to immunotherapy. We develop a transcriptional predictor of immunotherapy response and assess its prediction in genomic data from ~10,000 human tissues across 30 different cancer types to estimate the potential response to immunotherapy. The integrative analysis reveals two distinct tumor types: the mutator type is positively associated with potential response to immunotherapy, whereas the chromosome-instable type is negatively associated with it. We identify somatic mutations and copy number alterations significantly associated with potential response to immunotherapy, in particular treatment with anti-CTLA-4 antibody. Our findings suggest that tumors may evolve through two different paths that would lead to marked differences in immunotherapy response as well as different strategies for evading immune surveillance. Our analysis provides resources to facilitate the discovery of predictive biomarkers for immunotherapy that could be tested in clinical trials.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
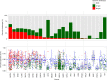

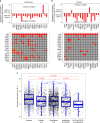
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

