Large-scale genomic analyses reveal the population structure and evolutionary trends of Streptococcus agalactiae strains in Brazilian fish farms
- PMID: 29051505
- PMCID: PMC5648781
- DOI: 10.1038/s41598-017-13228-z
Large-scale genomic analyses reveal the population structure and evolutionary trends of Streptococcus agalactiae strains in Brazilian fish farms
Abstract
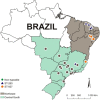
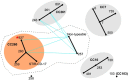
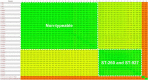
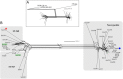
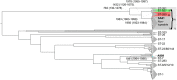
Streptococcus agalactiae is a major pathogen and a hindrance on tilapia farming worldwide. The aims of this work were to analyze the genomic evolution of Brazilian strains of S. agalactiae and to establish spatial and temporal relations between strains isolated from different outbreaks of streptococcosis. A total of 39 strains were obtained from outbreaks and their whole genomes were sequenced and annotated for comparative analysis of multilocus sequence typing, genomic similarity and whole genome multilocus sequence typing (wgMLST). The Brazilian strains presented two sequence types, including a newly described ST, and a non-typeable lineage. The use of wgMLST could differentiate each strain in a single clone and was used to establish temporal and geographical correlations among strains. Bayesian phylogenomic analysis suggests that the studied Brazilian population was co-introduced in the country with their host, approximately 60 years ago. Brazilian strains of S. agalactiae were shown to be heterogeneous in their genome sequences and were distributed in different regions of the country according to their genotype, which allowed the use of wgMLST analysis to track each outbreak event individually.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Streptococcus agalactiae Sequence Type 283 in Farmed Fish, Brazil.Emerg Infect Dis. 2019 Apr;25(4):776-779. doi: 10.3201/eid2504.180543. Emerg Infect Dis. 2019. PMID: 30882311 Free PMC article.
-
Microevolution of Streptococcus agalactiae ST-261 from Australia Indicates Dissemination via Imported Tilapia and Ongoing Adaptation to Marine Hosts or Environment.Appl Environ Microbiol. 2018 Aug 1;84(16):e00859-18. doi: 10.1128/AEM.00859-18. Print 2018 Aug 15. Appl Environ Microbiol. 2018. PMID: 29915111 Free PMC article.
-
Genomic comparison between pathogenic Streptococcus agalactiae isolated from Nile tilapia in Thailand and fish-derived ST7 strains.Infect Genet Evol. 2015 Dec;36:307-314. doi: 10.1016/j.meegid.2015.10.009. Epub 2015 Oct 9. Infect Genet Evol. 2015. PMID: 26455417
-
Comparative proteome analysis of two Streptococcus agalactiae strains from cultured tilapia with different virulence.Vet Microbiol. 2014 May 14;170(1-2):135-43. doi: 10.1016/j.vetmic.2014.01.033. Epub 2014 Feb 7. Vet Microbiol. 2014. PMID: 24594355
-
One hypervirulent clone, sequence type 283, accounts for a large proportion of invasive Streptococcus agalactiae isolated from humans and diseased tilapia in Southeast Asia.PLoS Negl Trop Dis. 2019 Jun 27;13(6):e0007421. doi: 10.1371/journal.pntd.0007421. eCollection 2019 Jun. PLoS Negl Trop Dis. 2019. PMID: 31246981 Free PMC article.
Cited by
-
In Vitro Pharmacodynamics and Bactericidal Mechanism of Fungal Defensin-Derived Peptides NZX and P2 against Streptococcus agalactiae.Microorganisms. 2022 Apr 22;10(5):881. doi: 10.3390/microorganisms10050881. Microorganisms. 2022. PMID: 35630326 Free PMC article.
-
Susceptibility of Tambaqui (Colossoma macropomum) to Nile Tilapia-Derived Streptococcus agalactiae and Francisella orientalis.Microorganisms. 2024 Nov 27;12(12):2440. doi: 10.3390/microorganisms12122440. Microorganisms. 2024. PMID: 39770643 Free PMC article.
-
Delineation of the pan-proteome of fish-pathogenic Streptococcus agalactiae strains using a label-free shotgun approach.BMC Genomics. 2019 Jan 7;20(1):11. doi: 10.1186/s12864-018-5423-1. BMC Genomics. 2019. PMID: 30616502 Free PMC article.
-
Serotype distribution, virulence factors, and antimicrobial resistance profiles of Streptococcus agalactiae (Group B Streptococcus) isolated from pregnant women in the Brazilian Amazon.BMC Microbiol. 2025 Jun 7;25(1):361. doi: 10.1186/s12866-025-04077-2. BMC Microbiol. 2025. PMID: 40481415 Free PMC article.
-
Transcriptome and Proteome of Fish-Pathogenic Streptococcus agalactiae Are Modulated by Temperature.Front Microbiol. 2018 Nov 2;9:2639. doi: 10.3389/fmicb.2018.02639. eCollection 2018. Front Microbiol. 2018. PMID: 30450092 Free PMC article.
References
-
- Evans JJ, et al. Characterization of β-haemolytic Group B Streptococcus agalactiae in cultured seabream, Sparus auratus L., and wild mullet, Liza klunzingeri (Day), in Kuwait. Journal of Fish Diseases. 2002;25:505–513. doi: 10.1046/j.1365-2761.2002.00392.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

