Retrotransposon Domestication and Control in Dictyostelium discoideum
- PMID: 29051748
- PMCID: PMC5633606
- DOI: 10.3389/fmicb.2017.01869
Retrotransposon Domestication and Control in Dictyostelium discoideum
Abstract
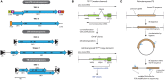
Transposable elements, identified in all eukaryotes, are mobile genetic units that can change their genomic position. Transposons usually employ an excision and reintegration mechanism, by which they change position, but not copy number. In contrast, retrotransposons amplify via RNA intermediates, increasing their genomic copy number. Hence, they represent a particular threat to the structural and informational integrity of the invaded genome. The social amoeba Dictyostelium discoideum, model organism of the evolutionary Amoebozoa supergroup, features a haploid, gene-dense genome that offers limited space for damage-free transposition. Several of its contemporary retrotransposons display intrinsic integration preferences, for example by inserting next to transfer RNA genes or other retroelements. Likely, any retrotransposons that invaded the genome of the amoeba in a non-directed manner were lost during evolution, as this would result in decreased fitness of the organism. Thus, the positional preference of the Dictyostelium retroelements might represent a domestication of the selfish elements. Likewise, the reduced danger of such domesticated transposable elements led to their accumulation, and they represent about 10% of the current genome of D. discoideum. To prevent the uncontrolled spreading of retrotransposons, the amoeba employs control mechanisms including RNA interference and heterochromatization. Here, we review TRE5-A, DIRS-1 and Skipper-1, as representatives of the three retrotransposon classes in D. discoideum, which make up 5.7% of the Dictyostelium genome. We compile open questions with respect to their mobility and cellular regulation, and suggest strategies, how these questions might be addressed experimentally.
Keywords: DIRS-1; RNA interference; Skipper-1; TRE5-A; retrovirus.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

