The plant circadian clock influences rhizosphere community structure and function
- PMID: 29053146
- PMCID: PMC5776454
- DOI: 10.1038/ismej.2017.172
The plant circadian clock influences rhizosphere community structure and function
Abstract
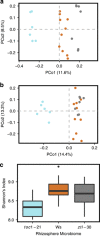
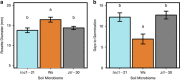
Plants alter chemical and physical properties of soil, and thereby influence rhizosphere microbial community structure. The structure of microbial communities may in turn affect plant performance. Yet, outside of simple systems with pairwise interacting partners, the plant genetic pathways that influence microbial community structure remain largely unknown, as are the performance feedbacks of microbial communities selected by the host plant genotype. We investigated the role of the plant circadian clock in shaping rhizosphere community structure and function. We performed 16S ribosomal RNA gene sequencing to characterize rhizosphere bacterial communities of Arabidopsis thaliana between day and night time points, and tested for differences in community structure between wild-type (Ws) vs clock mutant (toc1-21, ztl-30) genotypes. We then characterized microbial community function, by growing wild-type plants in soils with an overstory history of Ws, toc1-21 or ztl-30 and measuring plant performance. We observed that rhizosphere community structure varied between day and night time points, and clock misfunction significantly altered rhizosphere communities. Finally, wild-type plants germinated earlier and were larger when inoculated with soils having an overstory history of wild-type in comparison with clock mutant genotypes. Our findings suggest the circadian clock of the plant host influences rhizosphere community structure and function.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Badri DV, Vivanco JM. (2009). Regulation and function of root exudates. Plant Cell Environ 32: 666–681. - PubMed
-
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57: 233–266. - PubMed
-
- Bashan Y. (1998). Inoculants of plant growth-promoting bacteria for use in agriculture. Biotechnol Adv 16: 729–770.
-
- Berendsen RL, Pieterse CMJ, Bakker PAHM. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci 17: 478–486. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

